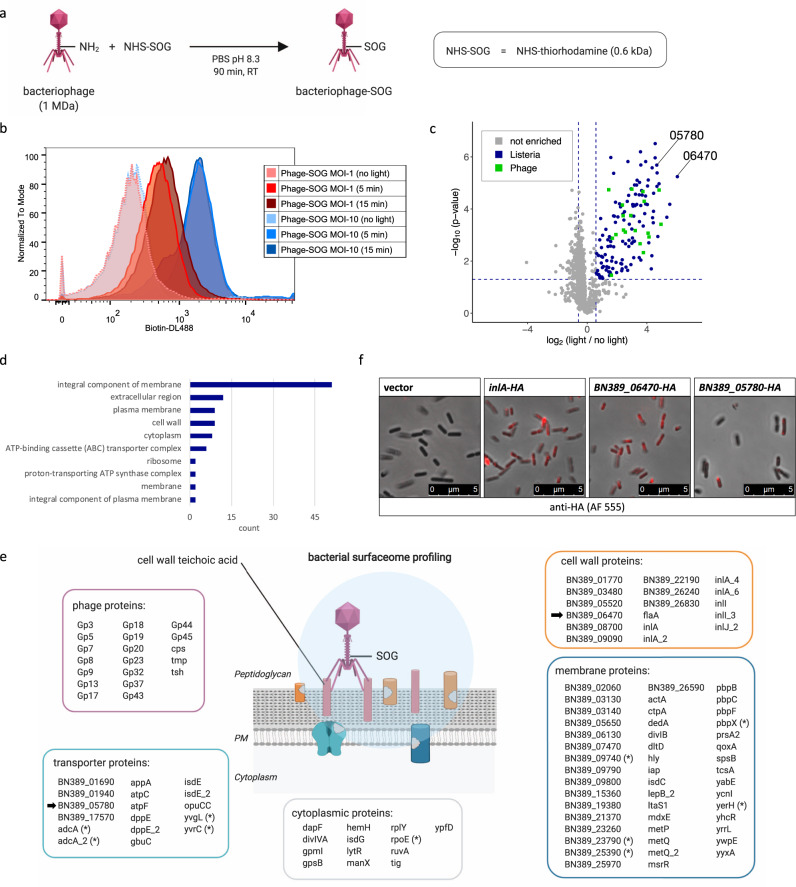
Fig. 5. Mapping virus-targeted surfaceomes on living hosts using bacteriophage-guided LUX-MS.
a Schematic of coupling the Listeria monocytogenes (Lm)-specific bacteriophage to the singlet oxygen generator (SOG) thiorhodamine. b Histogram plot showing light-dependent cell surface biotinylation of Lm cells after LUX-labeling with bacteriophage-SOG (>30,000 cells per condition). c Volcano plot showing relative abundance changes of LUX-MS quantified proteins from bacteriophage-SOG treated Lm with and without illumination for 15 min, tested using a two-sided Student’s t test. Blue and green dots represent significantly enriched Lm- or phage-derived proteins, respectively, whereas gray dots indicate non-enriched proteins. Candidates selected for microscopic characterization are highlighted. d GO-term enrichment analysis of significantly enriched Lm proteins. e Schematic representation of the spatial coverage of LUX-MS based on subcellular annotations of significantly enriched proteins. Candidates selected for microscopy are marked with arrows. f Immunofluorescence of the indicated Lm strains stained with an anti-HA monoclonal antibody conjugated with Alexa Fluor 555 (Red). Images are representative of three experiments. Source data are provided as a Source Data file and interactive volcano plots (Supplementary Data 5).