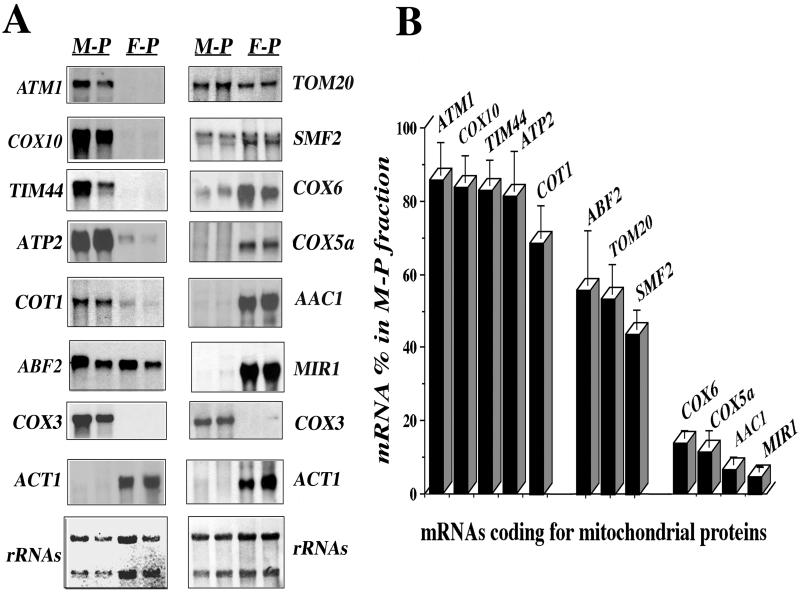
FIG. 1.
Asymmetric distribution of mRNAs coding for mitochondrial imported proteins among free and mitochondrion-bound polysomes. (A) CW04 cells were grown aerobically (rich galactose medium) and harvested in early log phase; free polysomes (F-P) and mitochondrion-bound polysomes (M-P) were prepared at pH 7.4 as described in Materials and Methods. Their RNA was extracted and subjected to Northern blot analysis, using probes for different genes encoding mitochondrial proteins. Cross-contamination of the fractions was checked with the ACT1 gene as a cytoplasmic protein control and the COX3 gene as a mitochondrial protein control. Results obtained for two individual RNA preparations are shown; at the bottom, methylene blue staining of the filters is shown. The approximate sizes measured for individual mRNAs were as follows: ATM1, 2.2 kb; COX10, 1.7 kb; TIM44, 1.5 kb; ATP2, 1.6 kb; COT1, 1.5 kb; ABF2, 0.6 kb; COX3, 3.7 kb; ACT1, 1.7 kb; TOM20, 0.8 kb; SMF2, 1.9 kb; COX6, 0.6 kb; COX5a, 0.6 kb; AAC1, 1.1 kb; and MIR1, 1.2 kb. (B) Densitometric analyses of the results obtained with 12 independent polysome preparations were performed. A signal obtained for an individual transcript in the RNA preparation from mitochondrion-bound polysomes was normalized with the COX3 mRNA signal. The normalization for the signal in the free-polysome fraction was performed with the ACT1 mRNA signal. For a given mRNA, addition of specific signals measured in mitochondrion-bound polysomes and in free polysomes after normalization was considered as 100%. The percentage of mRNA signal found in mitochondrion-bound polysomes is shown for the 12 genes examined.