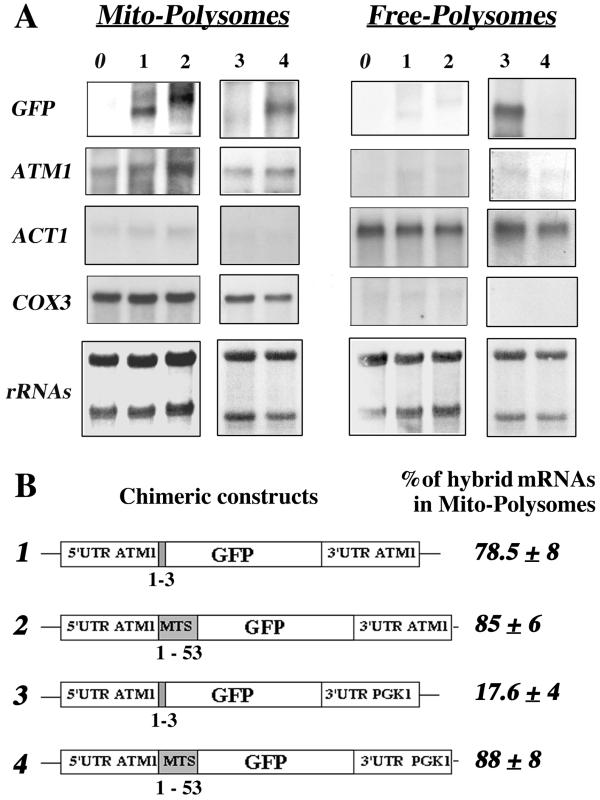
FIG. 4.
ATM1 sequences required for the asymmetric subcellular distribution of GFP hybrid mRNAs. (A) Wild-type CW04 cells (lane 0) or cells carrying the four plasmids (lanes 1 to 4) described in Fig. 3 C were grown until early log phase and harvested in order to purify free and mitochondrion-bound polysomes (see Materials and Methods). Then, 10 μg of RNA extracted from each polysomal fraction was separated on formaldehyde-agarose gels, subjected to Northern blot analysis, and hybridized successively with GFP, ATM1, ACT1, and COX3 probes. Methylene blue staining of the filters is shown at the bottom. Results obtained for RNA prepared from mitochondrion-bound polysomes (Mito-Polysomes) are shown in the left panel, and those obtained from free cytoplasmic polysomes (Free-Polysomes) are shown in the right panel. The autoradiograms represent exposure times of between 2 and 4 h at −80°C, with Amersham intensifying screens, for all the probes except ATM1, which required an exposure time of approximately 16 h. (B) Chimeric constructs are represented, as well as the percentage of the hybrid mRNA signal measured in polysomes bound to mitochondria, given as a mean of six independent experiments. In all cases, the presence of the ATM1 mts or its 3′UTR allowed the GFP hybrid mRNA to behave, with respect to its final subcellular localization, as the endogenous ATM1 mRNA.