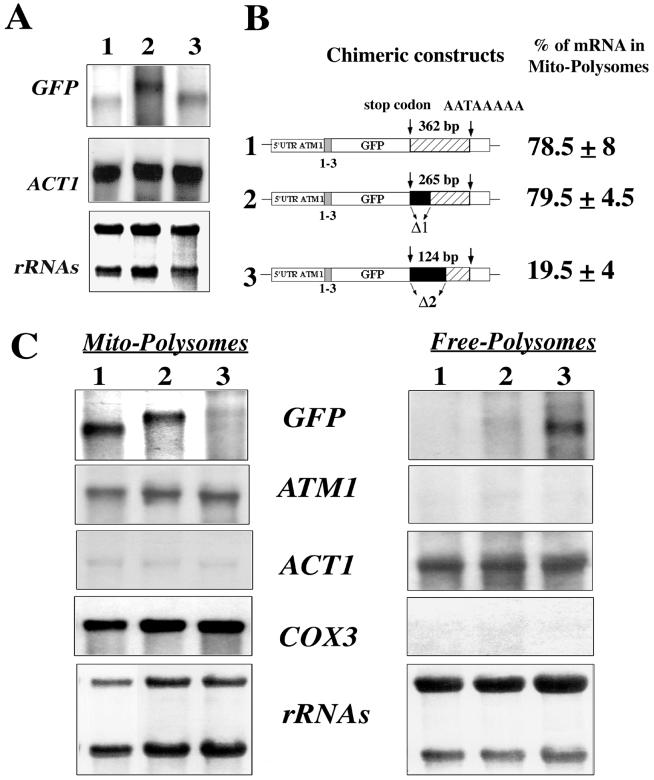
FIG. 6.
Minimal region within the 3′UTR of ATM1 required for the correct localization of GFP hybrid mRNAs. (A) Northern blots were performed using RNA extracted from whole cells carrying plasmids in which the regions between the stop codon and the canonical polyadenylation signal sequence AATAAA was shortened by 238 and 97 nucleotides, respectively. The steady-state levels of these individual mRNAs were quite similar in all of the cells tested. The approximate sizes for each hybrid mRNA were as follows: plasmid 1, 0.7 kb; plasmid 2, 1 kb; and plasmid 3, 0.8 kb. (B) Two plasmids were constructed in which deletions of the ATM1 3′UTR were performed. In plasmid 1 the complete 507 bp region of the 3′UTR was associated with the stop codon; the stop codon and the canonical polyadenylation signal sequence AATAAA are separated in this plasmid by 362 nucleotides. In plasmids 2 and 3, 97 and 238 nucleotides, respectively, shortened the region between the stop codon and the AATAAA signal. In all three chimeric constructs, the GFP is associated in frame with the first three amino acids of Atm1p. Calculations of the relative abundance of each transcript in mitochondrion-bound polysomes, for six independent experiments, were obtained after the normalization of each signal with the internal markers COX3 or ACT1. (C) Northern blots performed with RNAs purified from mitochondrion-bound polysomes (Mito-Polysomes) and free cytoplasmic polysomes (Free-Polysomes). Methylene blue staining of the filters is shown at the bottom. The autoradiograms represent exposures times of between 2 and 4 h at −80°C, with Amersham intensifying screens, for all of the probes except ATM1, which required an exposure time of approximately 16 h.