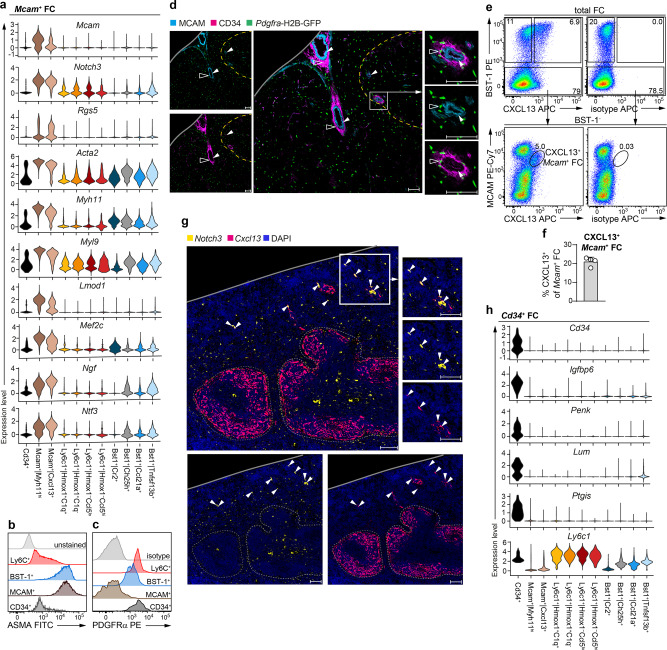
Fig. 3. Mcam+ FC and Cd34+ FC are distinct components of the splenic perivascular niche.
a Violin plots showing expression of marker genes that segregate Mcam+ FC (scaled total UMI counts per cell) across individual clusters. b, c Flow cytometric analysis of b ASMA or c PDGFRα expression by the indicated FC subsets. Representative stains from 2 independent experiments with a single biological replicate per experiment using pooled cell preparations from 2 spleens/replicate. d Confocal microscopy analysis of MCAM (light blue), CD34 (pink) on splenic sections from mice expressing H2B-GFP fusion gene (i.e. nucleus-confined GFP) from the endogenous Pdgfra locus (green). Exemplary Mcam+ FC (MCAM+GFP−) are indicated by filled arrowheads. Exemplary Cd34+ FC (CD34+GFP+) are indicated by empty arrowheads. Dashed yellow line, the white pulp/red pulp border. Solid grey line, the capsule. Scale bars are 50 μm. Representative stains from 3 spleens analysed in 2 independent experiments. e, f Flow cytometric validation of Mcam+|Cxcl13+ FC. e Representative stains. Numbers are percentage of cells in the indicated gates. f Percentage of CXCL13+ cells amongst Mcam+ FC. Data are pooled from 2 independent experiments and presented as arithmetic mean ± SD of n = 4 mice (depicted as symbols). g In situ RNA hybridization (RNAscope) for Cxcl13 (pink) and Notch3 (yellow). Sections were counterstained with DAPI (dark blue). Exemplary Mcam+|Cxcl13+ FC (Notch3+Cxcl13+ cells) are indicated by arrowheads. Dashed grey line, the white pulp/red pulp border. Solid grey line, the capsule. Scale bars are 100 μm. Representative stains from 2 spleens analysed in 2 independent experiments. h Violin plots showing expression of marker genes that segregate Cd34+ FC (scaled total UMI counts per cell) across individual clusters. a, h Violin plots were generated using default settings of the function VlnPlot() of the R package Seurat.