Abstract
Acute myeloid leukaemia (AML) is a neoplasm of immature myeloid cells characterized by various cytogenetic alterations. The present study showed that in addition to the FLT3-ITD and NPM1 mutation status, telomere length (TL) and telomerase reverse transcriptase (TERT) gene polymorphisms may affect risk and overall survival (OS) in AML. TL was longer in healthy controls than in AML patients and positively correlated with age in the patients, but not in healthy subjects. TL was found to be independently affected by the presence of the FLT3-ITD mutation. As for the TERT gene polymorphism, AML patients with the TERT rs2853669 CC genotype were characterized by significantly shorter OS than patients carrying the T allele. Another observation in our study is the difference in TL and OS in patients belonging to various risk stratification groups related to the FLT3-ITD and NPM1 mutation status. Patients with adverse risk classification (mutation in FLT3-ITD and lack of mutation in NPM1) presented with the shortest telomeres and significantly worse OS. In conclusion, OS of AML patients appears to be affected by TERT gene variability and TL in addition to other well-established factors such as age, WBC count, or FLT3-ITD and NPM1 mutation status.
Subject terms: Cancer genetics, Haematological cancer
Introduction
Acute myeloid leukaemia (AML) is a heterogeneous haematological malignancy, characterized by clonal expansion of abnormal immature leukaemic blasts1–3. Molecular changes in driver genes, polymorphic abnormalities and coexisting common mutational spectra occurring in AML are important prognostic and predictive markers in younger as well as in older AML patients4–6. Basic risk stratification and prognostic scoring of AML is based on the presence of mutation within nucleophosmin member 1 gene (NPM1) and/or signal transduction fms-like tyrosine kinase 3 (FLT3) gene, and groups patients into favourable, intermediate, and adverse risk categories7. There are many other gene mutations described in AML that are stratified according to the different functional pathways in which these genes are involved (e.g., oncogenes, transcription factors, tumour suppressors, epigenetic and chromatin modifying genes), most of which are important in diagnostics and modern therapy of AML patients8.
The FLT3-internal tandem duplication (FLT3-ITD) is the most common genetic alteration and is identified in approximately 25% of AML patients9,10. It leads to proliferative activation by continuous phosphorylation of the FLT3 receptor, and at the same time suppresses apoptosis. In clinical practice, FLT3-ITD mutations are independent markers of poor prognosis in cytogenetically normal AML10. Moreover, they are associated with an aggressive disease phenotype and shorter overall survival11. Another important gene abnormality identified in both young and older AML patients is the NPM1 mutation12. This mutation is found in almost one-third of newly diagnosed cases and leads to mislocalized NPM1 protein, found in the cytoplasm instead of the nucleolus13. In the absence of the FLT3-ITD alteration, NPM1 mutation is associated with a favourable prognosis and possibility for complete remission of the disease in AML patients14.
AML it the most common haematological neoplasm associated with short telomeres15. The occurrence of telomere shortening in leukaemias depends particularly on telomerase activity, telomerase reverse transcriptase catalytic subunit (TERT) expression, TERT promoter gene mutation (TERTp), and variability within the TERT gene16–19. Reduction of telomerase activity and extremely short telomeres induce chromosomal instability, causing bone marrow failure. TERT overexpression is observed in 80–95% of malignant cells and this dysregulation in the cancer cells can be explained by factors that lead to modification e.g. in the TERT promotor structure20.
A better understanding of the molecular mechanisms underlying AML led to development of drugs and new treatment strategies21. Standard clinical treatment of AML patients consists of high-intensity induction chemotherapy and/or haematopoietic stem cell transplantation22. However, many newly diagnosed AML patients do not qualify for intensive chemotherapy because of their age (> 75 years) or comorbidities23,24. Moreover, patients with FLT3 mutations are characterized with a much worse response to chemotherapy. Nowadays, therapies using various FLT3 tyrosine kinase inhibitors are applied in AML patients with FLT3 mutation24. Unfortunately, effective treatment of AML patients is challenging because of a very clonal heterogeneity of the disease and the occurrence of drug resistance.
In the present study we aimed to analyse AML patients in terms of the presence of FLT3-ITD and/or NPM1 gene mutations, telomere length and genetic variability within catalytic subunit of telomerase (TERT) in younger and old AML patients with respect to the clinical data, including overall survival (OS).
Results
Telomere length in patients and controls
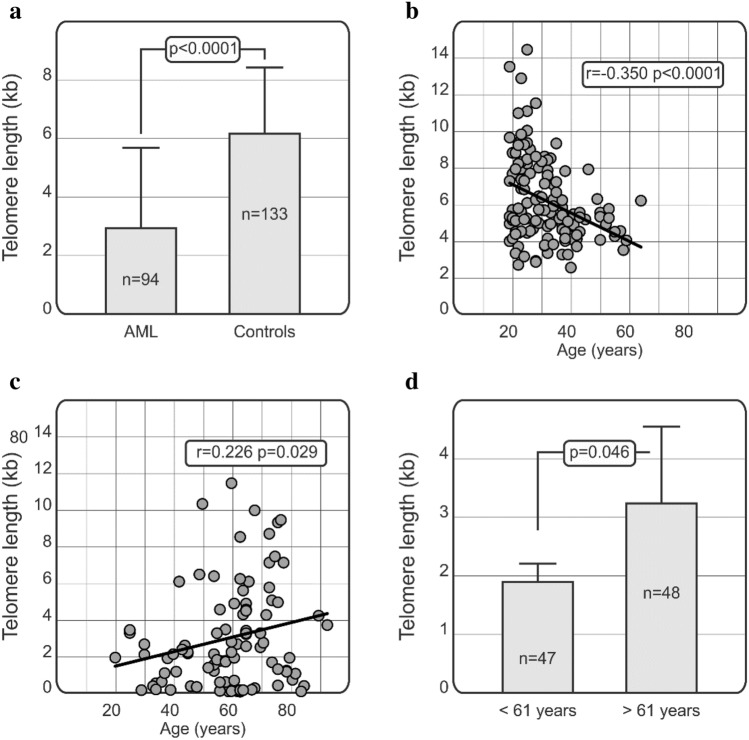
A significant difference (p < 0.0001) was observed between telomere length of healthy controls and AML patients (Fig. 1a), this was confirmed in an age-adjusted logistical regression analysis (p < 0.0001). In healthy controls, mean telomere length equalled 6.16 ± 2.27 kb (median: 5.56 kb, range from 2.58 kb to 14.43 kb) while in AML patients telomere length was shorter and equalled 2.94 ± 2.75 kb (median 2.09 kb, range from 0.02 kb to 11.48 kb). Moreover, telomere length declined with age in healthy subjects (r = − 0.350, p < 0.0001), while it increased in AML cases (r = 0.226, p = 0.029) (Fig. 1b,c).
Figure 1.
Comparison of telomere length in AML patients and controls (a) and relationships between age and telomere length in patients and controls. Telomere length correlates negatively with age in healthy controls (b) but not in AML patients (c). Statistical analysis was performed using Pearson correlation (PC) tests. Comparison of telomere length in AML patients below and above 61 years of age. Mann-Whitney U test was employed to assess the significance of differences in telomere length (d).
When the patients were subdivided with respect to the median age at diagnosis (61 years), it was observed that the older patients presented with longer telomeres as compared to younger patients (3.33 ± 2.84 vs 1.89 ± 3.73 kb, p = 0.046; Fig. 1d). There were 47 patients younger than 61 years and 48 older than 61 years.
Effect of the TERT gene polymorphism on overall survival
We did not observe any statistically significant differences in allele and genotype distribution between AML patients and healthy individuals for any of the TERT SNPs (rs2736100, rs2853669) studied (Table 1). Thus, in our AML patients, none of TERT (rs2736100, rs2853669) variants was found to affect the susceptibility of the disease.
Table 1.
Distribution of TERT genotypes in acute myeloid leukaemia (AML) patients and healthy individuals.
| AML patients (n = 91) | Healthy individuals (n = 133) | |
|---|---|---|
| TERT rs2736100 (intron 2) | ||
| CC | 21 (23.1%) | 31 (23.3%) |
| AC | 47 (51.6%) | 58 (43.6%) |
| AA | 23 (25.3%) | 44 (33.1%) |
| TERTp rs2853669 (promoter) | ||
| TT | 58 (63.7%) | 81 (61%) |
| CT | 24 (26.4%) | 43 (32%) |
| CC | 9 (9.9%) | 9 (7%) |
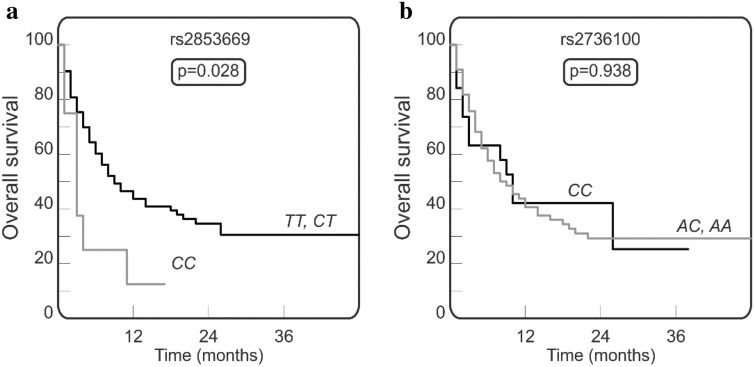
Employing Kaplan-Meier curves, we compared the overall survival of AML patients carrying various TERT genotypes. Patients with rs2853669 CC homozygous genotype presented with a shorter overall survival than patients having the rs2853669 T allele (carriers of TT or CT genotypes; p = 0.028; Fig. 2a). The most favourable effect on survival was observed for the rs2853669 CT heterozygosity (p = 0.089; not shown). As for the second TERT SNP (rs2736100) investigated, no significant association between the presence of any of its genetic variants and overall survival was observed (p = 0.961; Fig. 2b), although there was a trend towards better OS in elderly patients over 61 years old carrying the rs2736100 CC genotype (p = 0.051).
Figure 2.
Overall survival in AML patients carrying various genotypes of TERT rs2853669 (a) and TERT rs2736100 (b). The homozygous rs2853669 CC genotype is associated with shorter overall survival (a). The Gehan-Breslow-Wilcoxon test was used for statistical analysis.
FLT3-ITD and/or NPM1 mutation status in relation to telomere length, overall survival and other clinical parameters
As expected, analysis of the survival curves showed that younger patients (below median age of 61 years) lived longer than the older patients with a median overall survival of 12 and 5 months, respectively (p = 0.007).
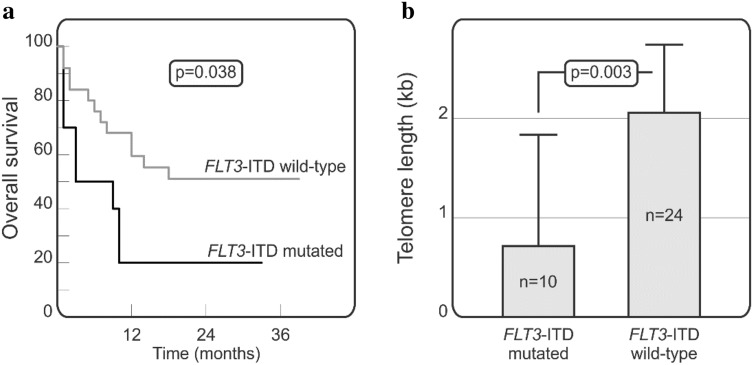
Interestingly, some additional associations related to the presence of the unfavourable FLT3-ITD mutation were noted in the group of patients below 61 years of age. We observed, that among AML patients below 61 years, those carrying the FLT3-ITD mutation had significantly lower median telomere length 0.72 ± 0.81 kb (range from 0.02 to 2.19 kb) when compared to FLT3-ITD wild type cases with median telomere length of 2.07 ± 2.30 kb (range from 0.1 to 10.35 kb) (p = 0.003; Fig. 3a). Moreover, the presence of FLT3-ITD mutation in this group of AML patients was found to be associated with significantly worse overall survival (p = 0.038; Fig. 3b). On the other hand, no significant relationships were observed for NPM1 mutation.
Figure 3.
The effect of the presence of FLT3-ITD mutation on telomere length and survival in patients below 61 years of age. Kaplan-Meier curves for overall survival in patients stratified with respect to the presence of FLT3-ITD mutation. Gehan-Breslow-Wilcoxon test was used for statistical analysis (a). Differences in median telomere length in AML patients having or lacking FLT3-ITD mutation assessed by Mann-Whitney U test (b).
Multivariate Cox regression analysis was employed to confirm independent associations of selected parameters with overall survival of AML patients. The following factors were considered: age, white blood cell (WBC) count, FLT3-ITD and NPM1 mutation status (mutated vs wild-type), telomere length as well as TERT rs2853669 (CT vs CC + TT) polymorphism. This analysis demonstrated that variability within the TERT gene and mutation status may influence overall survival. The analysis documented that rs2853669 heterozygosity (p = 0.0557) and the presence of NPM1 mutation (p = 0.0827) showed a positive association with overall survival, while higher WBC count (p = 0.0002) and more advanced age (p = 0.0448) showed an adverse effect (Table 2).
Table 2.
Multivariate analysis of factors potentially affecting overall survival in patients with AML.
| HR (95% CI) | p-value | |
|---|---|---|
| Age | 1.054 (1.001–1.109) | 0.0448 |
| WBC | 1.010 (1.004–1.014) | 0.0002 |
| FLT3-ITD, mutated vs wild-type | 2.854 (0.826–9.866) | 0.0975 |
| NPM1, mutated vs wild-type | 0.256 (0.055–1.193) | 0.0827 |
| Telomere length | 0.709 (0.455–1.105) | 0.1291 |
| TERT rs2853669 (CT vs CC + TT) | 0.324 (0.102–1.024) | 0.0557 |
HR Hazard ratio, CI confidence interval, WBC white blood cells count, FLT3-ITD internal tandem duplication of the FLT3 gene, NPM1 nucleophosmin member 1 gene.
Additional multivariate Cox regression analysis including age, WBC count, FLT3-ITD and NPM1 mutation status (mutated vs wild-type), as well as TERT rs2853669 (CC vs CT + TT) homozygosity showed that CC genotype was also associated with overall survival in AML patients (HR 8.066, p = 0.0230). As expected, the higher WBC count (HR 1.009, p < 0.0001) and advanced age (HR 1.053, p = 0.0352) showed a negative impact on overall survival. This analysis also confirmed that the presence of the FLT3-ITD (HR 3.518, p = 0.0410) and NPM1 (HR 0.272, p = 0.0680) mutations influence overall survival in an opposite way.
To assess whether the FLT3-ITD and/or NPM1 mutation status could act as an independent risk factor affecting telomere length in AML patients, a multiple linear regression model considering the presence of the FLT3-ITD and NPM1 mutation and both SNPs was employed. It was found that the presence of FLT3-ITD significantly affected telomere length that it was shorter in the FLT3-ITD positive cases (p = 0.002), while patients positive for the NPM1 mutations tended to have longer telomeres; p = 0.074). In this analysis, none of the two SNPs appeared to be an independent factor for telomere length.
As for the associations with other clinical parameters, we observed that patients positive for FLT3-ITD mutation showed increased lactate dehydrogenase (LDH) levels (with an average of 763.5 U/l; p = 0.048) and a tendency towards a higher WBC count (112.5 × 109/l; p = 0.072).
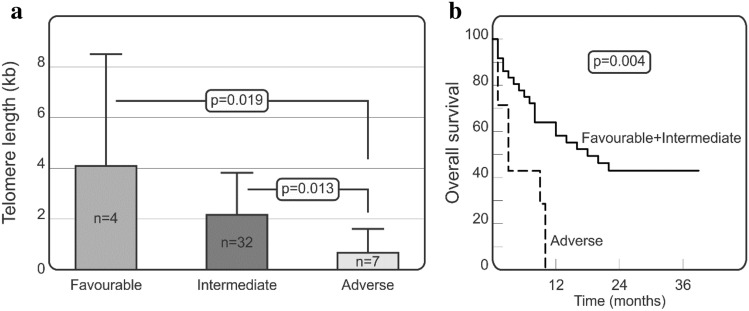
As the FLT3-ITD and NPM1 mutations are included, among other parameters, in the 2017 European LeukemiaNet (ELN) criteria7 of AML patients, we decided to check if telomere length and overall survival differ between patients in ELN risk groups (favourable, intermediate, adverse). As shown in Fig. 4a, AML patients with favourable risk classification characterized with longer telomeres as compared to AML patients with adverse risk. The mean telomere length equalled 4.08 ± 4.39 vs. 0.66 ± 0.94 kb, for patients with favourable and adverse risk respectively, p = 0.019; Fig. 4a). We also observed that patients with intermediate risk classification had longer telomeres as compared to the adverse group (2.15 ± 1.66 vs. 0.66 ± 0.94 kb, p = 0.013; Fig. 4a). Moreover, AML patients with adverse risk were characterized by worse overall survival as compared to patients of the other two risk groups (p = 0.002; Fig. 4b).
Figure 4.
Telomere length and overall survival of patients in various ELN risk groups. Kruskal–Wallis test with the Original FDR method of Benjamini and Hochberg were employed to assess the significance of differences in mean telomere length in AML patients with favourable, intermediate, and adverse risk stratification (a). Gehan–Breslow–Wilcoxon test for statistical analysis of overall survival curves in AML patients was performed (b).
Discussion
Acute myeloid leukaemia arises from the expansion of haematopoietic stem and progenitor cells which acquire numerous somatic mutations25. Approximately half of all AML patients are characterized by normal karyotype and have losses or duplications in terminal regions of chromosomes which may affect telomere stability26. Moreover, about 40–50 genes were found to harbour recurrent somatic mutations in various AML subtypes25. We hypothesised, that telomere length or polymorphisms within the TERT gene may also be related to disease risk, and survival.
In the present study we assessed telomere length in healthy blood donors and AML patients. We observed that AML patients were characterized by significantly shorter telomeres than healthy subjects. Similar relationships were shown by Aalbers et al. in the group of children with AML. They observed that telomere length in leukaemic cells was very short as compared to healthy control peripheral blood mononuclear cells16. Ventura Ferreira et al. also demonstrated that AML patients were characterized by significantly shorter telomeres than healthy controls27. Moreover, in our present study we have also noticed that telomere length decreases with age in healthy controls, but not in AML cases. Previously published data show inconsistent results. For example, a study by Menshawy et al. demonstrated a lack of correlation between age and telomere length in patients with AML28. On the contrary, Williams et al. reported a correlation between telomere length and age of diagnosis in AML patients29. Interestingly, both studies, our present one and that of Williams et al., report differences in telomere length between patients at different ages. We divided our AML patients into two groups, below and above 61 years of age (reflecting the median age in our group of patients). Our analysis showed that younger patients did have shorter telomere length as compared to older patients. Similar results were reported by Williams et al., who observed that younger AML patients (< 60 years old) had significantly shorter telomeres than patients at a more advanced age29.
In several studies, genetic variability within the TERT gene was analyzed in order to look for association with predisposition to the disease, its progression/outcome, or survival. We analyzed two TERT SNPs, one in the TERT promoter region (rs2853669, C/T) and the other located in intron 2 (rs2736100, C/A). We did observe some associations between TERT polymorphism and overall survival, although no significant relationship with the risk for the development of AML was found. In our study, patients carrying homozygous rs2853669 CC genotype were characterized with shorter overall survival than patients with T allele while the CT heterozygosity seemed to play more favourable role (see Fig. 3). Our results are consistent with those of Mosrati et al. in Swedish patients with AML. Additionally, Mosrati et al. found an interesting association between TERT rs2853669 CC homozygosity and increased expression of IL-6 and TNFα, cytokines known as markers for inflammation and cancer progression19. Furthermore, they reported that the rs2736100 SNP generated a modest risk for AML, although it had no effect on survival in their AML cohort. The latter observation confirms our results on the lack of association between rs2736100 and survival. On the other hand, it has been reported that in Chinese population, rs2736100 is associated with increased susceptibility to non-small cell lung cancer and myeloproliferative neoplasm30–32 as well as AML risk33. Tong et al. observed a higher frequency of the CC genotype and C allele of rs2736100 polymorphism in the Chinese AML patients. However, no significant differences were detected in either genotype or allele distributions between patients and control groups regarding the second SNP (rs2853669)33. The above observations may suggest that the effect of TERT SNPs may vary between patients from different populations and may be dependent on the broader genetic background of examined populations.
AML is the most common haematologic neoplasm that is associated with short telomeres. Watts et al. suggested that shorter telomere length leads to disconnection of proteins [e.g., telomeric repeat-binding factor 2 (TRF2)] from telomere complex. This biological process may be correlated with loss of major non-telomeric functions such as DNA damage and activation of important repair pathways. Therefore, it seems that telomere length could confer resistance to cytotoxic chemotherapy by affecting DNA repair mechanisms34.
Mutations in FLT3-ITD and NPM1 genes are frequently identified in AML, especially in patients with de novo AML and normal karyotype. Both of these genes are involved in important cellular processes, such as differentiation and apoptosis of haematopoietic progenitor cells. The occurrence of mutation in the NPM1 gene may be beneficial for the health and survival of patients2. On the other hand, excessive proliferation and survival of FLT3-ITD mutant cell clones have adverse effects for AML patients and is associated with poor prognosis35. In our present study, we found some differences between patients having and lacking the FLT3-ITD mutation in terms of telomere length and overall survival. Similar relationships were observed also in some previous studies16,29,34. Moreover, Molina Garay et al. demonstrated that patients with the FLT3-ITD mutation characterized with significantly shorter lifespan as compared to patients lacking the FLT3-ITD mutation or with a mutation in the tyrosine kinase domain (FLT3-TKD)36.
As for the NPM1 mutation, no association between NPM1 mutation status and telomere length was found either in our present work or in previous studies16,29. However, in our logistic regression model, NPM1 mutation showed a slight trend towards association with survival.
The novelty of our study is the observation regarding the differences between telomere length in patients belonging to various risk stratification groups (according to the 2017 European LeukemiaNet criteria by Döhner et al.7). AML patients carrying NPM1, but not FLT3-ITD mutation (favourable risk group) were characterized by longer telomeres as compared to AML patients of the adverse risk group (with FLT3-ITD mutation and NPM1 wild-type). Moreover, we observed that patients of the intermediate risk group (FLT3-ITD mutation and NPM1 mutation or FLT3-ITD wild-type and NPM1 wild-type) similarly had significantly longer telomeres in comparison to the adverse risk group of patients. Our AML patients from the adverse risk group were characterized by worse overall survival as compared to patients of the other two risk groups.
Logistic regression analysis showed that WBC count is a strong independent risk factor for survival and the presence of FLT3-ITD mutation was associated with higher WBC counts in our AML patients. The latter relationship between WBC count at diagnosis and FLT3-ITD mutation status was also reported in a meta-analysis published by Picharski et al.37.
In summary, outcomes of patients with the FLT3-ITD mutation were significantly worse than of those without this mutation, and higher WBC count was associated with poor prognosis, probably because of the presence of FLT3 mutation that is likely associated with higher WBC count. We also observed a correlation between LDH levels and FLT3-ITD mutation status. Patients positive for FLT3-ITD mutation showed higher levels of LDH, suggesting an unfavourable role of LDH in AML patients. In line with this observation, the study of Djunic et al. showed that serum LDH level was the most significant predictor of poor complete remission ratio in AML patients38.
In conclusion, overall survival of AML patients depends on various factors such as age, telomere length, mutation status, and TERT variability. The presence of FLT3-ITD and NPM1 mutations is used for estimating survival and response to treatment, although new prognostic genetic factors could be used to construct a more detailed risk stratification system. Adverse risk patients (positive for FLT3-ITD but negative for NPM1 mutation) need novel approaches to improve their overall survival and to get a better response to treatment. To clearly demonstrate the role and significance of telomere length in AML patients with normal karyotype, a clinical study involving a larger number of patients may be needed. It seems necessary to explore the new genetic and environmental factors that could be involved in leukaemogenesis.
Methods
Characteristics of the study groups
The study involved 95 Polish patients diagnosed with de novo AML with normal karyotype (57 males and 38 females, age range 20–93, median 61 years). Blood samples were collected at diagnosis after obtaining informed consent from patients. All methods were according to the Declaration of Helsinki. Approval of the Bioethical Committee of Wroclaw Medical University was obtained for the study (No. KB-368/2019). WBC count range was 0.7–510.5 × 109/l (median = 21.5 × 109/l). Risk stratification groups included 11% patients with favourable risk, 49% patients with intermediate risk and 40% patients with adverse risk according to 2017 European LeukemiaNet criteria7. There were 11 AML patients with FLT3-ITD mutation and 34 without it (n = 45). Additionally, there were 8 patients with NPM1 mutation and remaining 35 without it (n = 43). The median overall survival was 9 months (range 1–122 months). Additionally, 133 blood donors (84 males and 49 females, age range 19–64, median 30 years) served as a control group for the TERT polymorphisms and telomere length studies. Due to difference between patients and controls, an age adjusted logistic regression analysis was additionally performed.
DNA extraction
Genomic DNA was isolated from 10 mL of peripheral blood taken on EDTA using the Qiagen DNA Isolation Kit (Qiagen, Hilden, Germany) following the recommendation of the manufacturer. DNA concentration and purity were quantified on DeNovix (DeNovix Inc., USA). Isolated DNA was used to for TERT genotyping and assessment of the telomere length in AML patients.
Genotyping of TERT gene polymorphisms
The selection of investigated single nucleotide polymorphisms (SNPs) within the TERT gene was based on results from the SNP Function Prediction tool of the National Institute of Environmental Health Sciences (NCBI Database) website and other auxiliary databases (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html; https://www.ncbi.nlm.nih.gov/snp/; https://www.ensembl.org/index.html) the following criteria were used: minor allele frequency in Caucasians above 10%, change in RNA and/or amino acid chain, potential splicing site and/or miRNA binding site.
Based on the above criteria, 2 SNPs were selected for the study. TERT rs2736100 (G > T), located in intron 2, is a susceptibility factor for a variety of cancers and myeloproliferative neoplasms. The TERT rs2853669 (T > C) SNP located at − 245 bp (Ets2 binding site) in the promoter region, suppresses TERT expression and is associated with the enzymatic activity of telomerase. The TERT rs2736100 SNP was determined with the use of LightSNiP typing assays (TIB MOLBIOL, Berlin, Germany) while a TaqMan assay was employed for rs2853669 SNP genotyping (ThermoFisher Scientific, USA). Both assays are based on real-time polymerase chain reactions (PCR). Amplifications were performed on a LightCycler480 II Real-Time PCR system (Roche Diagnostics International AG, Rotkreuz, Switzerland) according to the recommendations of the manufacturer. The PCR conditions were as follows: 95 °C for 10 min followed by 45 cycles of: 95 °C for 10 s, 60 °C for 10 s and 72 °C for 15 s. PCR was followed by one cycle of: 95 °C for 30 s, 40 °C for 2 min, and gradual melting from 75 °C to 40 °C.
Quantification of telomere length
The average telomere length was measured in genomic DNA samples of 91 AML patients and 133 controls. DNA samples were diluted with nuclease-free water to reach a concentration of 5 ng/mL. Telomere length measurements were performed on a LightCycler480 II Real-Time PCR system (Roche Diagnostics International, Rotkreuz, Switzerland) using quantitative polymerase chain reaction (qPCR) assay kits (ScienCell’s Absolute Human Telomere Length Quantification qPCR Assay Kit [AHTLQ], Carlsbad, CA, USA), as previously described39. The PCR conditions were as follows: 95 °C for 10 min followed by 32 cycles of: 95 °C for 20 s, 52 °C for 20 s and 72 °C for 45 s. Data analysis was conducted according to manufacturer’s instruction. All reactions were run in three replicates.
Statistical analysis
The null hypothesis that there is no difference between allele and genotype frequencies between patients and controls was tested with the Fisher's exact test, calculated using the web‐based tool http://vassarstats.net/tab2x2.htm. Survival was assessed using the Gehan-Breslow-Wilcoxon test and Kaplan-Meier survival curves. The remaining statistical analyses of differences between groups were performed using one-way analysis of variance (ANOVA; to determine the significance of differences between the groups), and the resulting p-values were FDR-adjusted using the Benjamini and Hochberg method. For each experiment, data normality was verified with the Shapiro–Wilk test. Considering that distribution of some data deviated from normal distribution, the non-parametric U Mann-Whitney test was performed for comparison of telomere length. Statistical calculations were performed by GraphPad Prism software (GraphPad Software, La Jolla, CA, version 8.0.1) and Real Statistics Resource Pack for Microsoft Excel 2019 (version 16.0.10369.20032, Microsoft Corporation, Redmont, Washington, USA). RStudio (RStudio, PBC, Boston, Massachusetts, USA) was used for multiple linear regression model analyses and logistic regression model (Cox regression model) analyses. The r value for the correlation was determined using Pearson correlation (PC) tests. Probability (p) values < 0.05 were considered statistically significant, while those between 0.05 and 0.10 as indicative of a trend. Data in the figures are presented as mean + Standard Deviation (SD) or median + 95% Confidence Interval (CI).
Acknowledgements
This research was supported by the TARGETTELO project No. STRATEGMED3/306853 from the National Centre for Research and Development, Warsaw, Poland. The authors are grateful to dr Tomasz Kubik for his help and assistance.
Author contributions
M.D.: conceptualization, methodology, formal analysis, investigation, writing—original draft, writing—review & editing. B.W.: conceptualization, methodology, formal analysis, writing—original draft, writing—review & editing. A.B.: resources, data curation (patient recruitment, sample collection, provision of clinical data). P.Ł.: formal analysis, writing—original draft, writing—review & editing. G.M.: resources, data curation (patient recruitment, sample collection, provision of clinical data). K.B.K.: conceptualization, methodology, formal analysis, writing—original draft, writing—review & editing, supervision, project administration. All authors have read and agreed to the published version of the manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N. Engl. J. Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Mahmood R, Altaf C, Malik HS, Khan SA. Clinico-Haematologic association and prognostic relevance of NPM1 and FLT3-ITD mutations in acute Myeloid Leukaemia. Pak. J. Med. Sci. 2019;35:23–28. doi: 10.12669/pjms.35.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martignoles JA, Delhommeau F, Hirsch P. Genetic hierarchy of acute myeloid leukemia: From clonal hematopoiesis to molecular residual disease. Int. J. Mol. Sci. 2018;19:3850. doi: 10.3390/ijms19123850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullinger L, Döhner K, Döhner H. Genomics of acute myeloid leukemia diagnosis and pathways. J. Clin. Oncol. 2017;35:934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 6.Tyner JW, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kishtagari A, Levine RL, Viny AD. Driver mutations in acute myeloid leukemia. Curr. Opin. Hematol. 2020;27:49–57. doi: 10.1097/MOH.0000000000000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuffa E, et al. Revealing very small FLT3 ITD mutated clones by ultra-deep sequencing analysis has important clinical implications in AML patients. Oncotarget. 2015;6:31284–31294. doi: 10.18632/oncotarget.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaguchi M, et al. Prognostic impact of low allelic ratio FLT3-ITD and NPM1 mutation in acute myeloid leukemia. Blood Adv. 2018;2:2744–2754. doi: 10.1182/bloodadvances.2018020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levis MJ, et al. A next-generation sequencing-based assay for minimal residual disease assessment in AML patients with FLT3-ITD mutations. Blood Adv. 2018;2:825–831. doi: 10.1182/bloodadvances.2018015925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kayser S, Levis MJ. Clinical implications of molecular markers in acute myeloid leukemia. Eur. J. Haematol. 2019;102:20–35. doi: 10.1111/ejh.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunetti L, Gundry MC, Goodell MA. New insights into the biology of acute myeloid leukemia with mutated NPM1. Int. J. Hematol. 2019;110:150–160. doi: 10.1007/s12185-018-02578-7. [DOI] [PubMed] [Google Scholar]
- 14.Pastore F, et al. Combined molecular and clinical prognostic index for relapse and survival in cytogenetically normal acute myeloid leukemia. J. Clin. Oncol. 2014;32:1586–1594. doi: 10.1200/JCO.2013.52.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nogueira BMD, et al. Telomere length and hematological disorders: A review. In Vivo. 2020;34:3093–3101. doi: 10.21873/invivo.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aalbers AM, et al. Telomere length and telomerase complex mutations in pediatric acute myeloid leukemia. Leukemia. 2013;27:1786–1789. doi: 10.1038/leu.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lansdorp PM. Maintenance of telomere length in AML. Blood Adv. 2017;1:2467–2472. doi: 10.1182/bloodadvances.2017012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonseka LN, Tirado CA. Telomerase in acute myeloid leukemia: A molecular update on diagnosis, prognosis, and treatment. J. Assoc. Genet. Technol. 2016;42:105–110. [PubMed] [Google Scholar]
- 19.Mosrati MA, et al. Association between TERT promoter polymorphisms and acute myeloid leukemia risk and prognosis. Oncotarget. 2015;6:25109–25120. doi: 10.18632/oncotarget.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, et al. Epigenetic landscape of the TERT promoter: A potential biomarker for high risk AML/MDS. Br. J. Haematol. 2016;175:427–439. doi: 10.1111/bjh.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang YW, et al. Cytogenetics and mutations could predict outcome in relapsed and refractory acute myeloid leukemia patients receiving BCL-2 inhibitor venetoclax. Ann. Hematol. 2020;99:501–511. doi: 10.1007/s00277-020-03911-z. [DOI] [PubMed] [Google Scholar]
- 22.Patel KK, et al. Cost-effectiveness of azacitidine and venetoclax in unfit patients with previously untreated acute myeloid leukemia. Blood Adv. 2021;5:994–1002. doi: 10.1182/bloodadvances.2020003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morsia E, et al. Venetoclax and hypomethylating agents in acute myeloid leukemia: Mayo Clinic series on 86 patients. Am. J. Hematol. 2020;95:1511–1521. doi: 10.1002/ajh.25978. [DOI] [PubMed] [Google Scholar]
- 24.Perl AE, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N. Engl. J. Med. 2019;381:1728–1740. doi: 10.1056/NEJMoa1902688. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. Ley TJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olbertova H, Plevova K, Stranska K, Pospisilova S. Telomere dynamics in adult hematological malignancies. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2019;163:1–7. doi: 10.5507/bp.2018.084. [DOI] [PubMed] [Google Scholar]
- 27.Ventura Ferreira MS, et al. Evidence for a pre-existing telomere deficit in non-clonal hematopoietic stem cells in patients with acute myeloid leukemia. Ann. Hematol. 2017;96:1457–1461. doi: 10.1007/s00277-017-3049-z. [DOI] [PubMed] [Google Scholar]
- 28.Menshawy NE, Ashwah SE, Ebrahim MA. Short dysfunctional telomere is highly predictive of dismal outcome in MDS but not in AML patients. Int. J. Hematol. Oncol. Stem Cell Res. 2020;14:188–199. doi: 10.18502/ijhoscr.v14i3.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams J, et al. Telomere length is an independent prognostic marker in MDS but not in de novo AML. Br. J. Haematol. 2017;178:240–249. doi: 10.1111/bjh.14666. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum. Mol. Genet. 2014;23:6616–6633. doi: 10.1093/hmg/ddu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahlström J, et al. TERT rs2736100 genotypes are associated with differential risk of myeloproliferative neoplasms in Swedish and Chinese male patient populations. Ann. Hematol. 2016;95:1825–1832. doi: 10.1007/s00277-016-2787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong T, Luo M, Liu Q. The TERT rs2736100 polymorphism and susceptibility to myeloproliferative neoplasms: A systematic review and meta-analysis. Genet. Test Mol. Biomark. 2020;24:181–187. doi: 10.1089/gtmb.2019.0277. [DOI] [PubMed] [Google Scholar]
- 33.Tong Y, et al. Association between TERT gene polymorphisms and acute myeloid leukemia susceptibility in a Chinese population: A case-control study. Cancer Cell Int. 2020;20:313. doi: 10.1186/s12935-020-01335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watts JM, et al. Telomere length and associations with somatic mutations and clinical outcomes in acute myeloid leukemia. Leuk. Res. 2016;49:62–65. doi: 10.1016/j.leukres.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia. 2019;33:299–312. doi: 10.1038/s41375-018-0357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molina Garay C, et al. Profiling FLT3 mutations in mexican acute myeloid leukemia pediatric patients: Impact on overall survival. Front. Pediatr. 2020;8:586. doi: 10.3389/fped.2020.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picharski GL, et al. The impact of Flt3 gene mutations in acute promyelocytic leukemia: A meta-analysis. Cancers. 2019;11:1311. doi: 10.3390/cancers11091311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djunic I, et al. Prognostic risk score for the survival of elderly patients with acute myeloid leukaemia comprising comorbidities. Med. Oncol. 2013;30:394. doi: 10.1007/s12032-012-0394-6. [DOI] [PubMed] [Google Scholar]
- 39.Dratwa M, et al. Heterogeneity of telomerase reverse transcriptase mutation and expression, telomerase activity and telomere length across human cancer cell lines cultured in vitro. Exp. Cell Res. 2020;396:112298. doi: 10.1016/j.yexcr.2020.112298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.