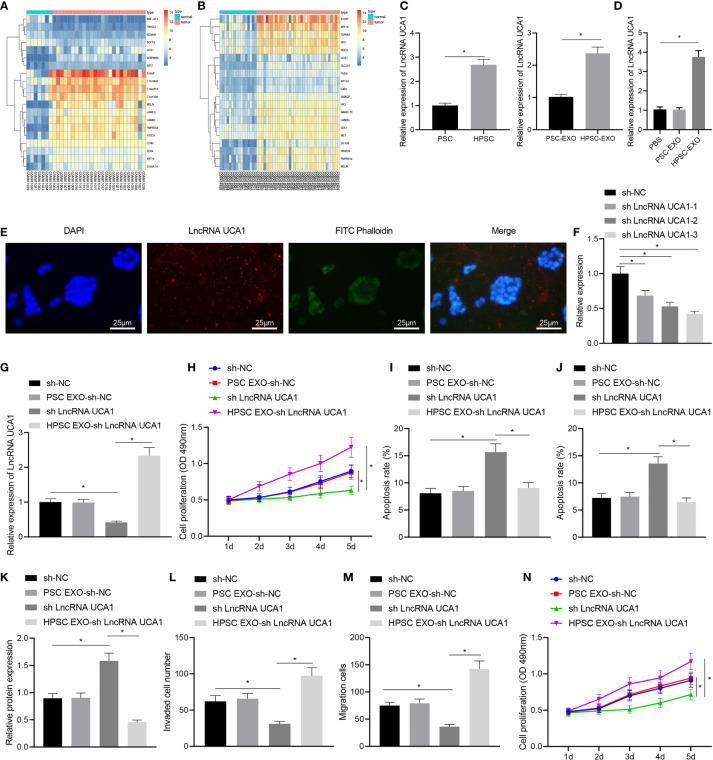
Figure 3.
HPSC-EXO transfer lncRNA UCA1 to pancreatic cancer cells, where lncRNA UCA1 stimulates the malignant phenotypes of pancreatic cancer cells and their resistance to Gem. (A), A heatmap of the expression of the first 10 differentially expressed genes in pancreatic cancer samples in the GSE32676 dataset. (B), A heatmap of the expression of the first 10 differentially expressed genes in pancreatic cancer samples in the GSE16515 dataset. (C), Expression of lncRNA UCA1 in PSCs and HPSCs as well as their derived exosomes was detected by qRT-PCR. (D), Expression of lncRNA UCA1 in PANC-1 cells treated with PSC-EXO and HPSC-EXO was detected by qRT-PCR. (E), Co-localization of lncRNA UCA1 and exosomes in PANC-1 cells was observed by a fluorescence microscope. (F), Transfection efficiency of three lncRNA UCA1 shRNA sequences in PANC-1 cells was detected by qRT-PCR. (G), Expression of lncRNA UCA1 in the PANC-1 cells treated with sh-lncRNA UCA1 or combined with HPSC-EXO was detected by qRT-PCR. (H), Viability of PANC-1 cells treated with sh-lncRNA UCA1 or combined with HPSC-EXO was detected by MTT assay. (I), Apoptosis of PANC-1 cells treated with sh-lncRNA UCA1 or combined with HPSC-EXO was detected by flow cytometry. (J), Apoptosis of PANC-1 cells treated with sh-lncRNA UCA1 or combined with HPSC-EXO was detected by TUNEL staining. (K), Cleaved caspase 3 protein expression in the PANC-1 cells treated with sh-lncRNA UCA1 or combined with HPSC-EXO was detected by Western blot. (L), Migration of PANC-1 cells treated with sh-lncRNA UCA1 or combined with HPSC-EXO was detected by Transwell assay. (M), Invasion of PANC-1 cells treated with sh-lncRNA UCA1 or combined with HPSC-EXO was detected by Transwell assay. (N), Gem sensitivity of PANC-1 cells treated with sh-lncRNA UCA1 or combined with HPSC-EXO was detected by MTT assay. *p < 0.05. Measurement data were expressed as mean ± standard deviation. The data between two groups were compared using the independent sample t test. The data comparison among multiple groups was performed using one-way ANOVA, followed by the Tukey’s post hoc test. The data at different time points were compared by repeated measures ANOVA, followed by Bonferroni’s post hoc test. The cell experiment was repeated three times.