Figure 1.
Reliability of murine brain organoid model
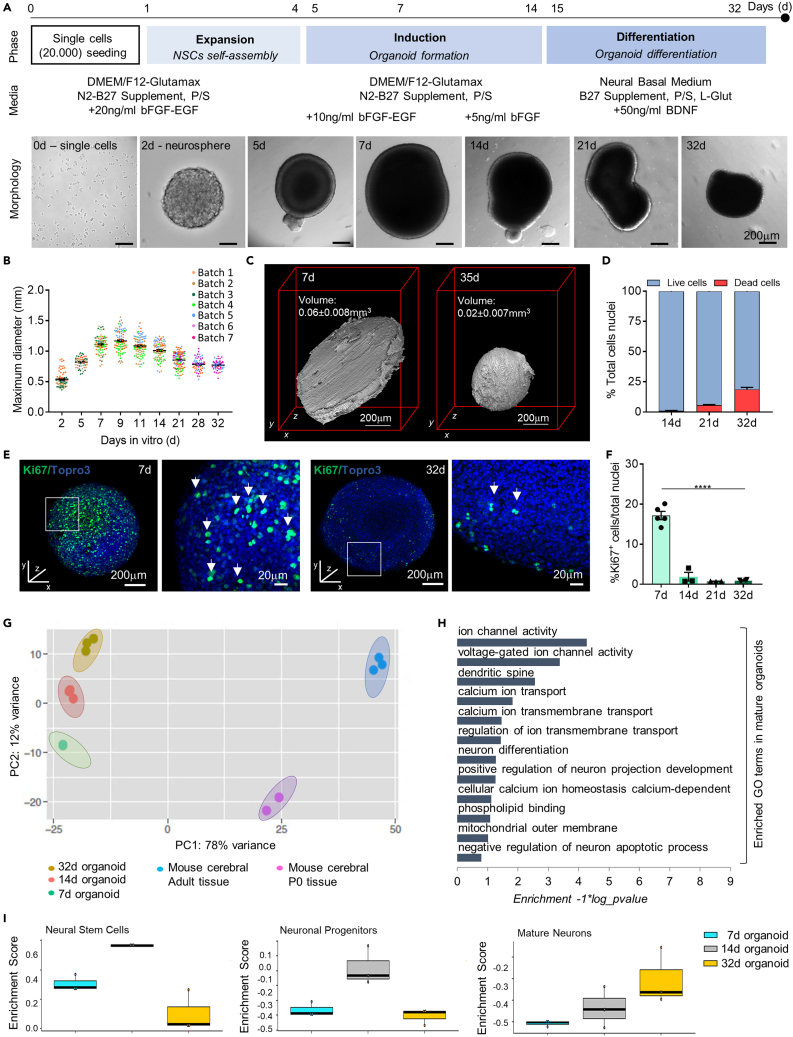
(A) Schematic representation of the three-phase organoid generation protocol. After single cells (20000 cells/well) seeding on a 24-well plate (day 0), the protocol starts with the initial Expansion Phase that lasts up to day 4: mouse NSCs proliferation and neurosphere formation in culture medium enriched with bFGF and EGF (20ng/ml). Induction Phase from day 5 to day 14: early organoid formation in medium supplemented with gradual decrease of bFGF and EGF (day 5 to day 6: 10ng/ml bFGF and EGF; day 7 to day 14: 5ng/ml bFGF). Differentiation Phase from day 15 to day 32: organoids maturation in differentiation medium (BDNF 50ng/ml) up to day 32. In all the three phases, organoids were maintained on an orbital shaker (dynamic culture). Pictures in (A) are representative brightfield images of single NSCs, a neurosphere and organoids at different stages of the protocol. At day 5 the early formed organoids show the presence of brightened translucent surface tissue, whereas, the central core is quite dark. Late organoids develop gradual dark and compact tissue morphology and structural remodeling.
(B) Graph representing the growth curve of murine brain organoids. An exponential growth is observed up to day 11 whereas the maximum diameter progressively diminishes as the differentiation proceeds. Data are expressed as mean ± SEM of the maximum diameter (n>30). Organoids belonging to different SGZ-NSCs batches were reported in different colors.
(C) Optical projection tomography (OPT) reconstruction of representative organoids at day 7 and day 35 showing the 3D structure of the in vitro generated organoid. The reported organoids volume was calculated for n = 2 organoids at 7d and n = 3 organoids at 35d based on the 360-degree OPT reconstructions using Avizo. Data are expressed as mean ± SEM.
(D) Graph representing the number of live and dead cells expressed in % of total cells number. The analysis was performed on n = 3 organoids/time point. Dead cells were identified as propidium iodide positive cells while the number of live cells was obtained by subtracting dead cells from the total nuclei.
(E) Representative confocal immunofluorescence maximum intensity projections of z stack images of whole mount murine brain organoids at 7 and 32 days in culture, showing the proliferative marker Ki67 (in green). Total nuclei are visible in blue (TOPRO-3). White boxes indicate a magnification of a region of the whole mount organoids, presented as single plane image. White arrows show Ki67+ cells.
(F) Graph representing the percentage of Ki67+ cells over total cell nuclei at different time points. Data are expressed as mean ± SEM. Analysis was performed on n = 3 different organoids and at least on 3 entire sections for each organoid. p < 0.05 was considered statistically significant. Statistical differences between marker expression at different time points were calculated by ordinary one-way ANOVA followed by Tukey's multiple comparison test. ∗∗∗∗p < 0.0001.
(G) Principal components analysis (PCA) of RNAseq data from organoid samples at different stages of differentiation (7 days: n = 3; 14 days: n = 3; 32 days: n = 3) and from murine cerebral tissues (P0: n = 2; adult: n = 3). PCA shows organoids and tissues cluster according to their different maturation phases.
(H) Gene ontology (GO) analysis for up-regulated genes in mature (32 days) versus early (7 days) organoids. Based on global transcriptomic analysis, GO analysis was performed in order to identify GO terms enriched in the up-regulated genes in mature compared to early organoids and to classify them based on molecular function, biological process and cellular components.
(I) Box plots of GSVA RNAseq data analysis generated evaluating specific gene lists for neural stem cells, neuronal progenitors and mature neurons in 7d, 14d and 32d organoids showing upregulation of genes related to neuronal maturation in late organoids. The Enrichment Score represents the amount to which the genes in the specific gene lists are over-represented.