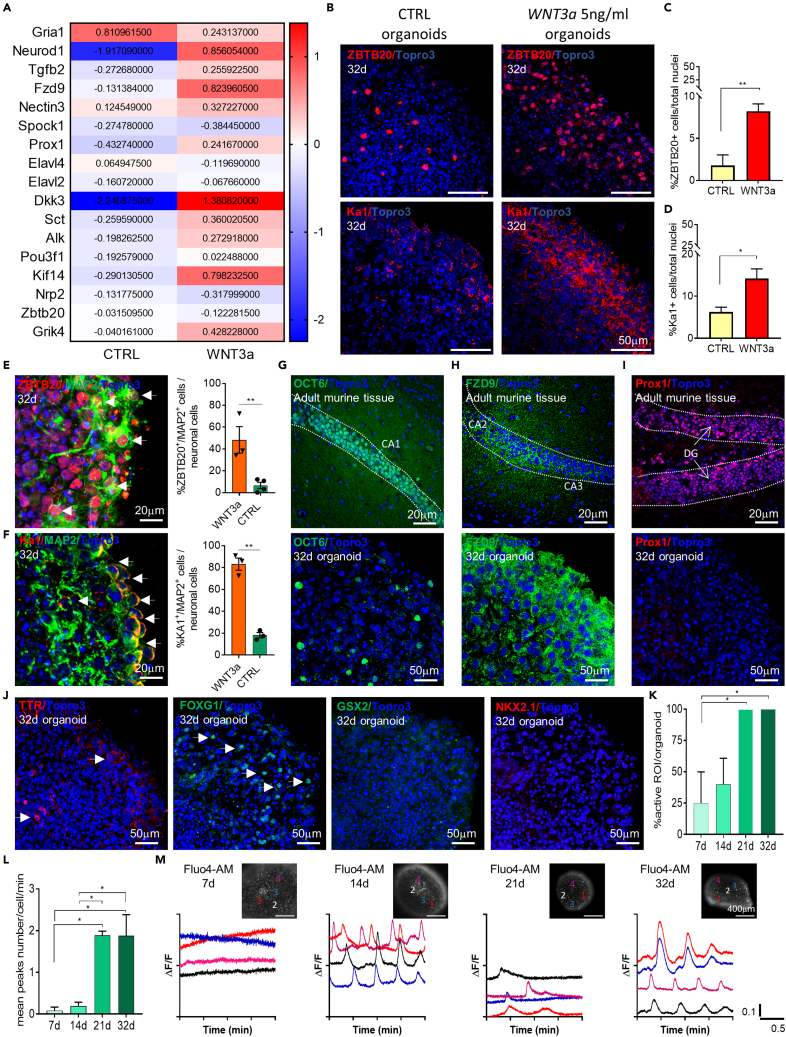
Figure 7.
Murine brain organoids develop CA3 hippocampal-specific neurons after WNT3a supplementation
(A) Heatmap showing the log2 fold change of genes related to hippocampal signature in control (CTRL) and WNT3a-treated (WNT3a) mature organoids.
(B) Representative confocal immunofluorescence images of sliced NSCs-derived control (CTRL) against WNT3a-treated organoids, at 32 d, showing cells positive for the pan-hippocampal ZBTB20 and the CA3-specific KA1 markers.
(C and D) Graphs showing the percentage of ZBTB20 (C) and Ka1 (D) positive cells over total cell nuclei at 32d in control and WNT3a-treated organoids. Analysis was performed on n≥3 different organoids and at least on 3 entire sections for each organoid. Data in all graphs are expressed as mean ± SEM. Statistical differences between experimental conditions were evaluated by two-tailed unpaired t test for two datasets (CTRL vs WNT3a) for both markers. ∗∗p<0.01; ∗p <0.05.
(E) Representative confocal immunofluorescence images of sliced WNT3a-treated organoids (32d), showing double-positive cells (highlighted by white arrows) for ZBTB20 and MAP2 mature neuronal markers. Graph on the right showing the percentage of ZBTB20+/MAP2+ cells over total neuronal cells in WNT3a and CTRL organoids at 32d. Data are expressed as mean ± SEM. Statistical differences were evaluated by two-tailed unpaired t test for two datasets (CTRL vs WNT3a). ∗∗p<0.01.
(F) Representative confocal immunofluorescence images of sliced WNT3a-treated organoids (32d), showing double-positive cells (highlighted by white arrows) for Ka1 and MAP2 mature neuronal markers. Graph on the right showing the percentage of Ka1+/MAP2+ cells over total neuronal cells in WNT3a and CTRL organoids at 32d. Data are expressed as mean ± SEM. Statistical differences were evaluated by two-tailed unpaired t test for two datasets (CTRL vs WNT3a). ∗∗p<0.01.
(G–I) Representative confocal immunofluorescence images of sliced adult (8 weeks) murine brain tissue and mature (32d) murine brain organoids showing the expression of OCT6 (G), FZD9 (H) and Prox1 (I) as CA1, CA2 and DG hippocampal markers, respectively. White insets represent a sketch of coronal mouse brain sections while red boxes highlight the brain region of the murine tissue staining.
(J) Representative confocal immunofluorescence images of mature (32d) murine WNT3a brain organoids showing the low or no expression of TTR-choroid plexus, FOXG1-cortical, GSX2 and NKX2.1 ganglionic eminences markers. All the pictures in B, E-F, G-J are maximum intensity projections of z stack images.
(K) Graph showing the percentage of active cells within the organoid at different time points (n≥3 organoids/time point; n = 10 cells selected in each organoid). Data in all graphs are shown as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. ∗p< 0.05.
(L) Graph showing the average number of the peaks counted for each cell per minute at different time points (n≥3 organoids/time point; n = 10 cells selected in each organoid). Data in all graphs are shown as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. ∗p< 0.05.
(M) Time-lapse Fluo4-AM calcium imaging on whole mount WNT3a-treated organoids at different time points, paired to representative graphs of calcium flux. Note the increasing neural activity over time as well as a major peak synchronization.