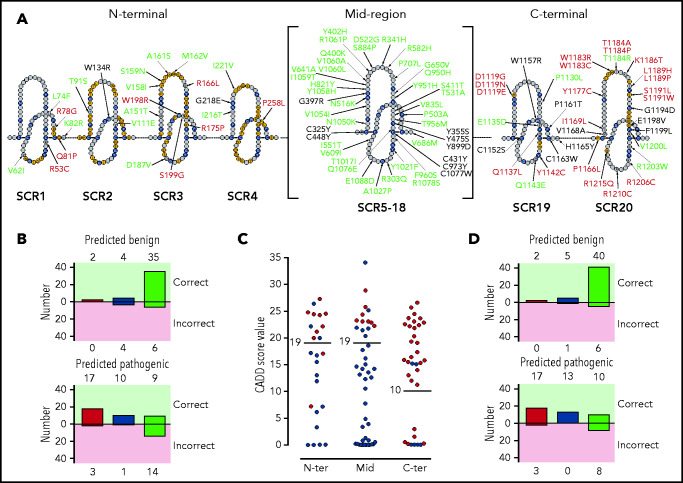
Figure 3.
Distribution of the tested FH variants in the FH sequence. (A) Schematic representation of the amino acid sequence of the 20 FH SCRs. Amino acids corresponding to the SCR consensus sequence are depicted in black, those functionally relevant are in yellow, and all other amino acids are in gray. FH variant classification codes are based on functional assay results: benign (green), nonexpressed (black), and functionally impaired (red). (B) Breakdown of CADD computational predictions using a default score value cutoff of 15: benign predictions (upper panel) and pathogenic predictions (lower panel). Residue types from left to right are “SCR consensus,” “functionally relevant,” and “other.” (C) Distribution of FH classified experimentally as pathogenic (red dots) or benign (green dots) according to CADD scores. Horizontal lines indicate the selected cutoff for each FH region. (D) Breakdown of CADD computational predictions using adjusted score value cutoff according to functional domain: benign predictions (upper panel) and pathogenic predictions (lower panel). Residue types from left to right are “SCR consensus,” “functionally relevant,” and “other.”