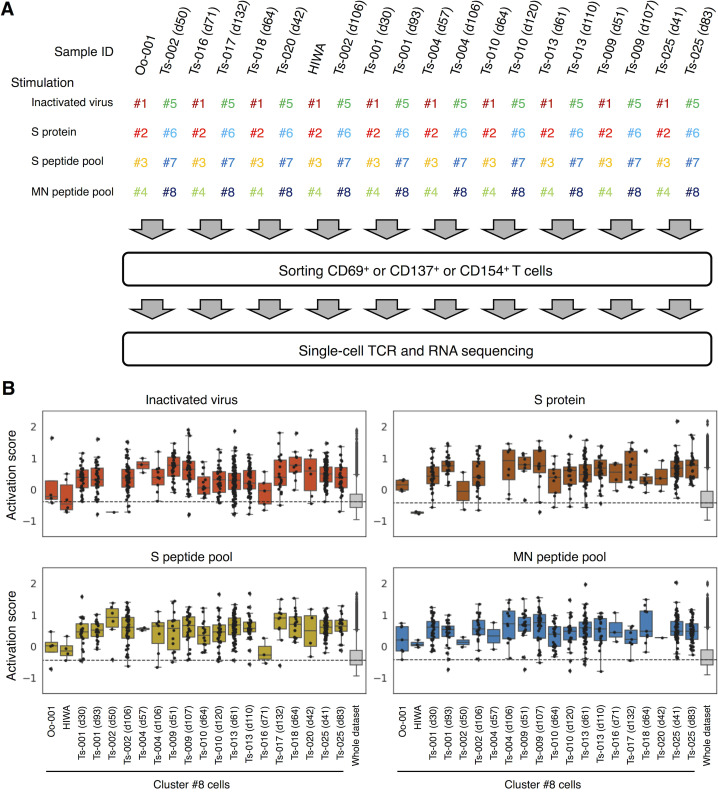
Figure S1.
Single-cell–based analyses of SARS-CoV-2–responsive T cells. PBMCs from healthy donors and convalescent COVID-19 patients were stimulated with SARS-CoV-2–derived antigens as indicated for 20 h before antigen-responding T cells were sorted and analyzed by single-cell TCR- and RNA-seq. (A) A workflow of single-cell–based analyses. Antigen-stimulated PBMCs were stained with eight different hashtag antibodies (#1–8), anti-CD3, and the indicated activation marker antibodies. Hashtagged cells from two patients were pooled. Antigen-responding (CD69+ or CD137+ or CD154+) CD3+ T cells were sorted and immediately subjected to single-cell TCR- and RNA-seq. Each collection day after diagnosis is shown after the donor identifier (ID). (B) Activation score of each single cell was calculated based on the expression of activation genes (CD69, TNFRSF9, TNFRSF4, and CD40LG).