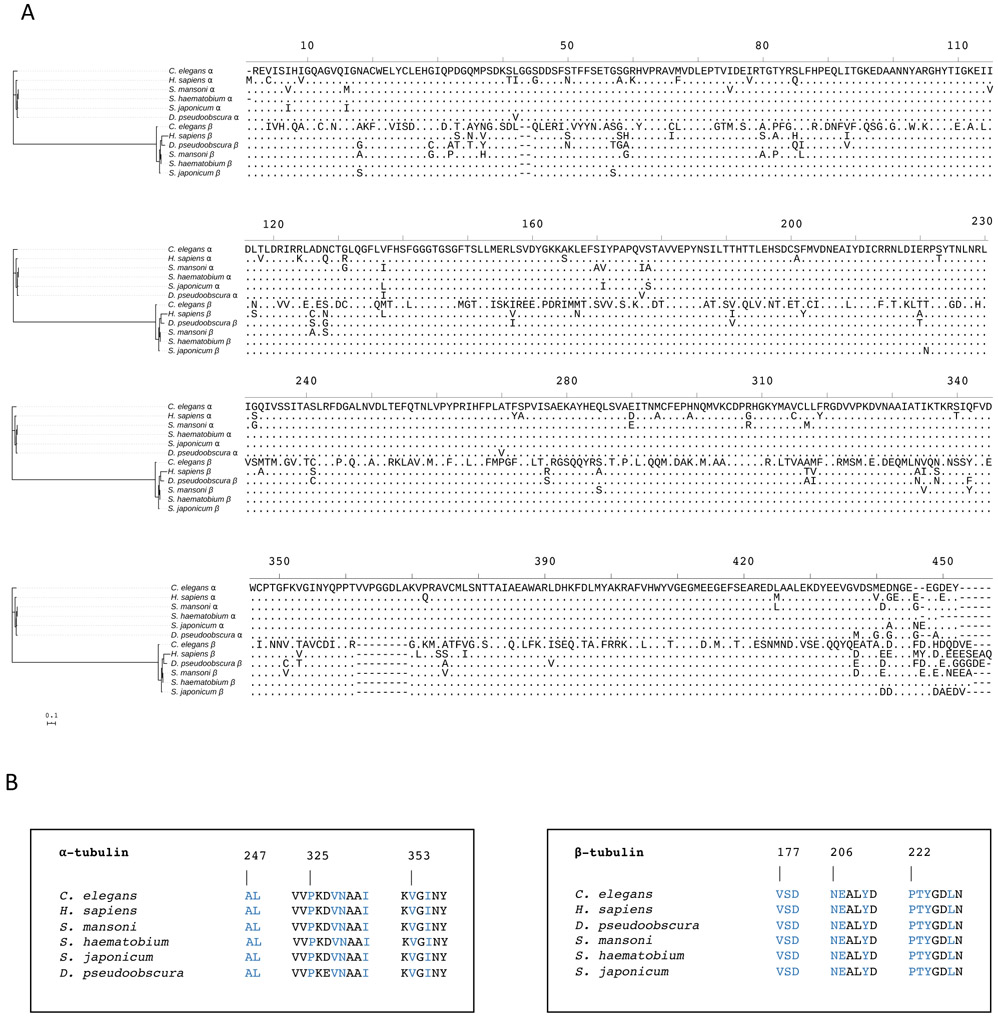
Figure 1. Phylogenetic comparison of α- and β-tubulins from key eukaryotic species.
(A) Full-length alignment of α- and β-tubulin amino acid sequences from S. mansoni, S. haematobium, S. japonicum, Drosophila pseudoobscura, Caenorhabditis elegans and Homo sapiens. (B) α- and β-tubulin amino acid residues in the proximity of the vinblastine binding site, which is at the interface between the β1 and α2-tubulin subunits. Amino acid residues in contact with a triazolopyrimidine (compound 1 in Ref. 23) in the 5NJH crystal structure23 are highlighted in blue. The alignment of full-length tubulin sequences was built using MUSCLE25, 26 and served as the input for the construction of a maximum likelihood phylogenetic tree using IQ-TREE2.27 The interactive tree of life program (iTOL)28 was used to visualise the phylogenetic tree and sequence alignment. The scale bar represents an approximate value for the amino acid substitution rate.