Figure 3: Transgenesis in Hofstenia enables a vast array of experimental tools for studying regeneration and stem cell biology.
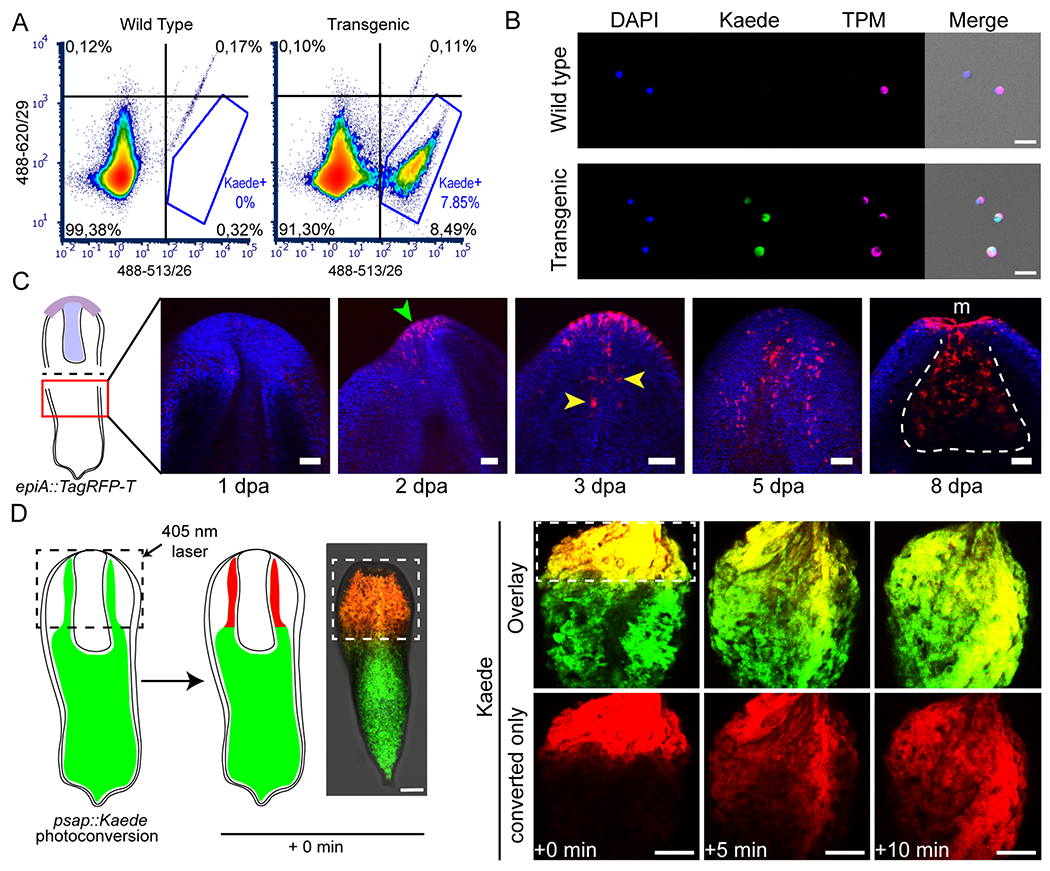
(A, B) Isolation of specific live cell populations using FACS on tnn::Kaede transgenic animals; (A) Fluorescence plots showing cell density for Kaede fluorescence (quadrant and Kaede gate percentages are calculated over a total of 76880 and 87157 singlets for wild type and transgenic samples, respectively); (B) anti-TPM immunocytochemistry on FACS-isolated cells originating from the whole cell (wild type), and the Kaede+ (transgenic) populations. Scale bars, 20 μm. FACS plot is representative of 3 biological replicates; immunocytochemistry was performed on two technical replicates of tnn::Kaede dissociated cells. (C) Regeneration of anterior structures following worm bisection (see sketch on the left) in epiA::TagRFP-T transgenics. Regeneration time points are indicated under each image, as days post amputation (dpa). Green and yellow arrowheads point at de novo expression of TagRFP-T in epidermal and pharyngeal cells, respectively. White dotted line delineates the regenerated pharynx, m, mouth. Scale bars, 50 μm. Six biological replicates were used for each time point (see figure S3). (D) Photoconversion of Kaede-expressing cells in live psap::Kaede animals (dotted squares outline the photoconverted area); left, schematic of the experiment; right, quick expansion of the photoconverted Kaede area (red) over time in a live worm (as observed in 3 biological replicates). Time elapsed since photoconversion is indicated at the bottom. Scale bars, 100 μm. See also figures S1 and S3.
