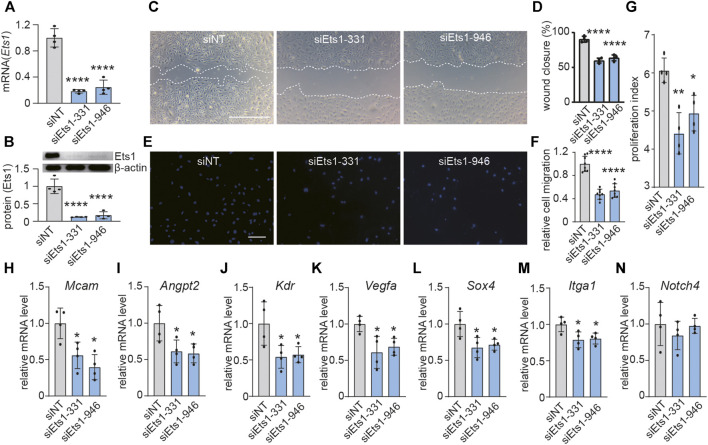
FIGURE 3.
Ets1 knockdown inhibits expression of genes associated with vascular abnormality. (A) Ets1 mRNA expression determined by qPCR in bEND.3 cells transfected with siNT (control siRNA) or siEts1 (mean + SD of four independent experiments). (B) Ets1 protein expression by western blot in bEND.3 cells trandfected with siNT or siEts1 (Top). Western blot quantification was performed with software Image J (bottom) (mean + SD of four independent experiments). (C, D) Micrographs (C) and quantification (D) of control or Ets1 silenced bEND.3 cell migration in scratch wound assays (bar: 200 μm) (n = 4/group). (E, F) Micrographs (E) and quantification (F) of control or Ets1 silenced bEend.3 cell migration in transwell migration assay (bar: 100 μm) (n = 8/group). (G) Quantification of control or Ets1 silenced bEend.3 cells in proliferation assay. (H–N) Quantification of mRNA expression of vascular abnormality associated genes including Mcam (H), Angpt2 (I), Kdr (J), Vegfa (K), Sox4 (L), Itga1 (M) and Notch4 (N) in control or Ets1 silenced cells (n = 4/group). Data represent the mean ± SD, One-way ANOVA with Tukey’s test, *p < 0.05, **p < 0.01, ****p < 0.0001.