Figure 1. WR and GxxxG motifs are not sufficient for ERAD-M retrotranslocation.
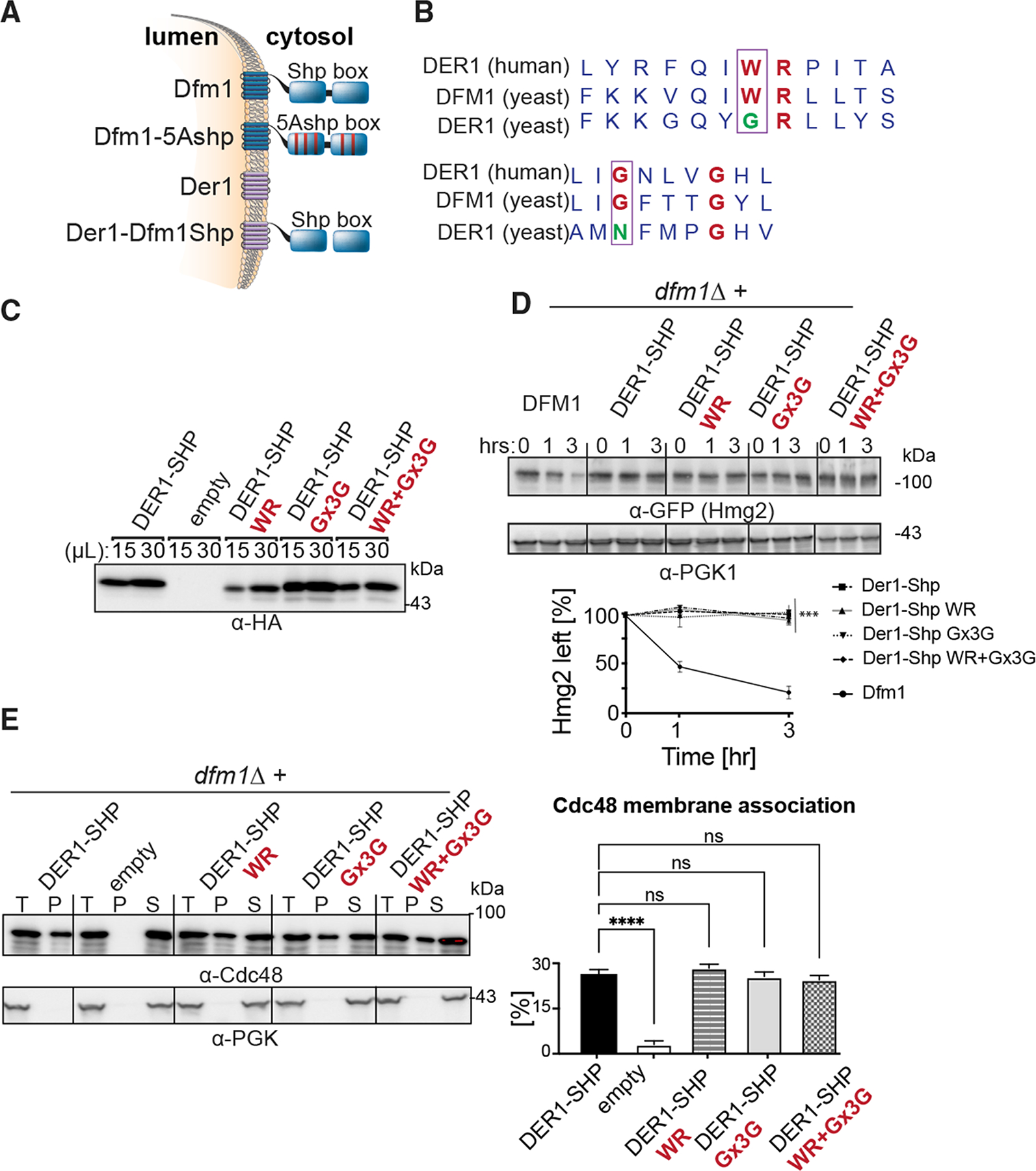
(A) Depiction of Dfm1, Der1, and Der1-Shp. Dfm1 and Der1 are ER-localized membrane proteins with six transmembrane domains.
(B) Alignment of WR and GxxxG motif from H. sapiens Derlin-1 and S. cerevisiae Der1 and Dfm1.
(C) Expression levels of Der1-Shp variants are measured by loading increasing amounts of lysates (15 and 30 μL) on SDS-PAGE followed by immunoblotting with α-HA.
(D) In the indicated strains, degradation of Hmg2-GFP was measured by CHX-chase assay. Cells were analyzed by SDS-PAGE and immunoblotted for Hmg2-GFP with α-GFP. Band intensities were normalized to PGK1 loading control and quantified by ImageJ. t = 0 was taken as 100%, and data are represented as mean ± SEM from n = 3 biological replicates, ***p < 0.001, repeated-measures ANOVA.
(E) Total cell lysates (T) from the indicated strains were separated into soluble cytosolic fraction (S) and pellet microsomal fraction (P) upon centrifugation at 14,000 × g. Each fraction was analyzed by SDS-PAGE and immunoblotted for Cdc48 with α-Cdc48 and Pgk1 with α-Pgk1. The graph shows the quantification of Cdc48 in the pellet fractions of the respective cells as measured from ImageJ. Data are represented as percentage of Cdc48 that is bound to pellet fraction and is shown as mean ± SEM from n = 3 biological replicates, ****p < 0.0001, one-way ANOVA.
