Figure 4. Dfm1’s WR motif and SHP box are required for interaction with membrane substrates.
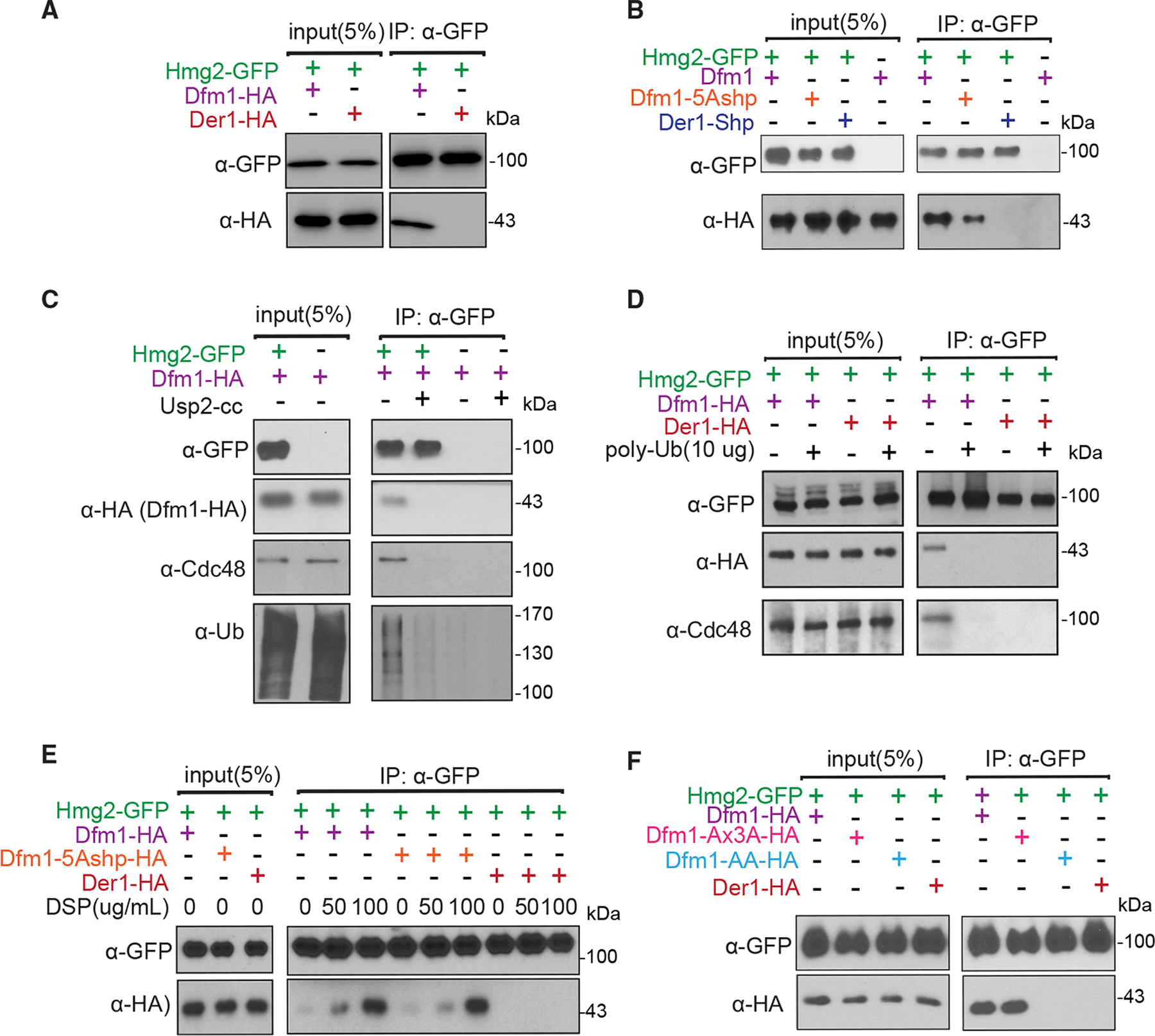
(A) Hmg2-GFP and Dfm1-HA binding were analyzed by coIP. As a control for specificity, cells expressing Der1-HA were used.
(B) Same as (A), except binding of Hmg2-GFP to Dfm1 variants, Dfm1-5Ashp and Der1-Shp, was analyzed.
(C) Dfm1-Cdc48 complex interacts directly with the polyubiquitin chain of Hmg2. Microsomes isolated from indicated strains were treated with Usp2Core, and Hmg2-GFP was immunoprecipitated, resolved on 8% SDS-PAGE, and immunoblotted for ubiquitin with α-Ub, Hmg2-GFP with α-GFP, Cdc48 with α-Cdc48, and Dfm1 with α-HA.
(D) Addition of Lys48-linked polyubiquitin chains disrupts binding of Hmg2 to Dfm1-Cdc48. Hmg2-GFP, Dfm1-HA, and Cdc48 binding were analyzed by coIP in the presence of an increasing amount of Lys48-linked polyubiquitin chains (2, 5, and 10 μg). As a negative control, strains not expressing Hmg2-GFP were used.
(E) Crosslinking analysis of Hmg2-GFP and Dfm1-5Ashp. Microsomes were harvested from DSP-treated strains and subjected to immunoprecipitation of Hmg2-GFP with GFP Trap, followed by immunoblotting for Dfm1-5Ashp with anti-HA and Hmg2 with anti-GFP.
(F) Same as (A), except binding to Hmg2-GFP was analyzed with Dfm1 variants: Dfm1-AA and Dfm1-Ax3A.
