Figure 5. Dfm1 L1 residues are required for binding to integral membrane substrates.
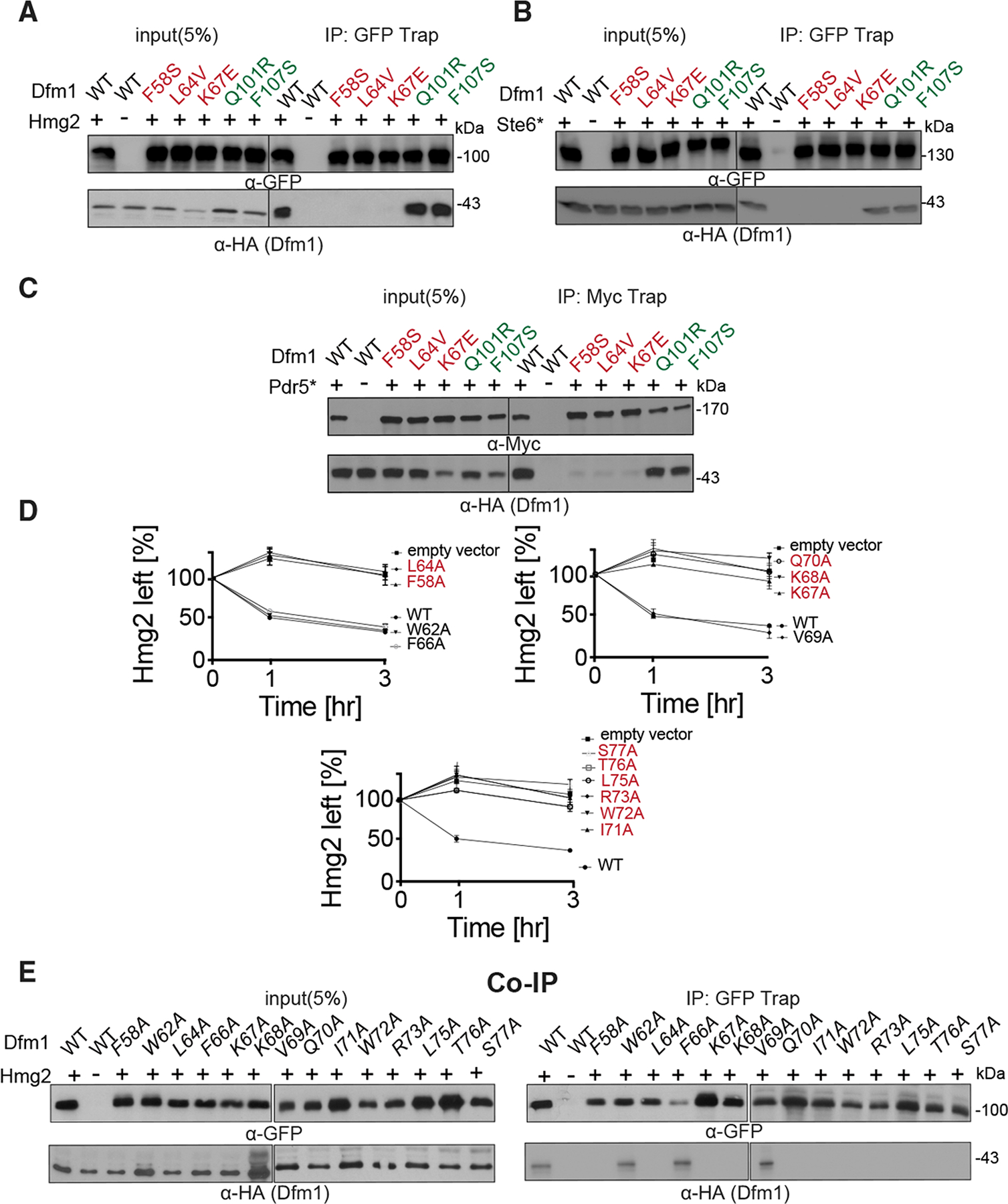
(A) Hmg2-GFP binding to retrotranslocation-deficient Dfm1 mutants was analyzed by coIP. As negative control, cells not expressing Hmg2-GFP were used.
(B) Same as (A), except Ste6* binding to retrotranslocation-deficient Dfm1 mutants was analyzed.
(C) Same as (A), except Pdr5* binding to retrotranslocation-deficient Dfm1 mutants was analyzed.
(D) Ala mutant scanning unveils additional L1 Dfm1 mutants required for ERAD. Strains were grown to log phase and subjected to CHX-chase. Hmg2-GFP levels were analyzed at the indicated times using flow cytometry. Histograms of 10,000 cells are shown, with the number of cells versus GFP fluorescence. Data from each time point are represented as mean ± SEM from n = 3 biological replicates.
(E) Same as (A), except Hmg2-GFP binding to L1 Dfm1 mutants generated by Ala mutant scanning were analyzed by coIP.
