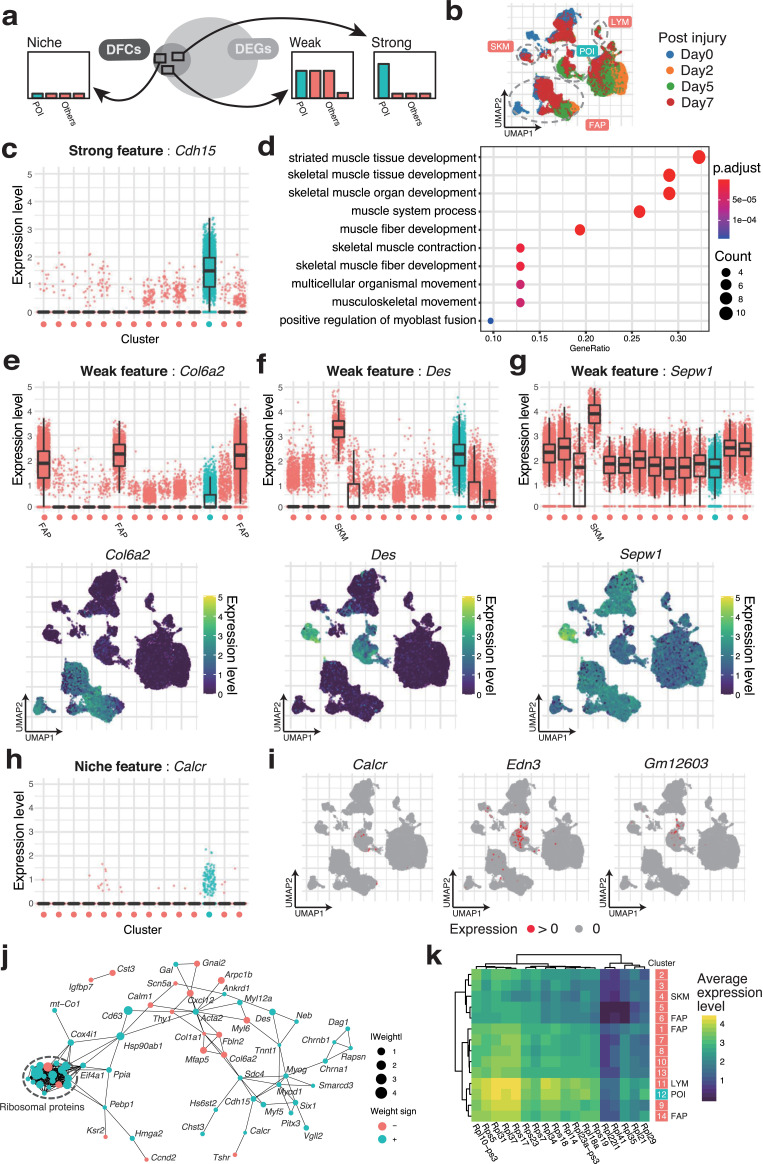
Fig 3. Biological significance of genes in DFC revealed by the discriminative ability.
(a) According to the specificity of the expression, the genes in DFC are classified into three groups. The three groups are named Strong (specific to 1–2 clusters), Weak (>2), and Niche features (none of them). (b) The data are from samples collected at 0, 2, 5, and 7 days after skeletal muscle injury. In addition, the clusters of fibro/adipogenic progenitors (FAPs), mature skeletal muscle (SKM), and lymphocytes (LYM) are shown. (c) Genes specifically expressed in the POI are assigned to the Strong features. For the Strong feature Cdh15, its expression level for each cluster shown in Fig 2B is plotted. The medians, 25th/75th percentiles, and 1.5 interquartile range (IQR) are employed to draw the box plots. (d) The Strong features contain many genes that act as markers of skeletal muscle. The results of GO enrichment analysis for the Strong features. GOs are ordered by the proportion of their inclusion in the Strong features. (e) Genes expressed in some clusters are assigned to the Weak features. For the Weak feature Col6a2, its expression level is plotted in the upper panel, and the single-cell expression level visualized by UMAP is plotted in the lower panel. (f, g) The expression levels of Sepw1 and Des, two of the Weak features, are plotted as (e). (h) Genes with low expression levels that are expressed in a minor subpopulation of the POI are assigned to the Niche features. For the Niche feature Calcr, its expression level is plotted as (c). (i) Cells expressing Niche features (Calcr, Edn3, and Gm12603) are highlighted. (j) DFC has the property of capturing interrelated genes. STRING is used to connect related DFC. (k) Ribosomal proteins are a notablef example of Weak features in DFC that are difficult to interpret in the context of binary combinations. Eighteen ribosomal protein genes in DFC are averaged in each cluster as a heat map.