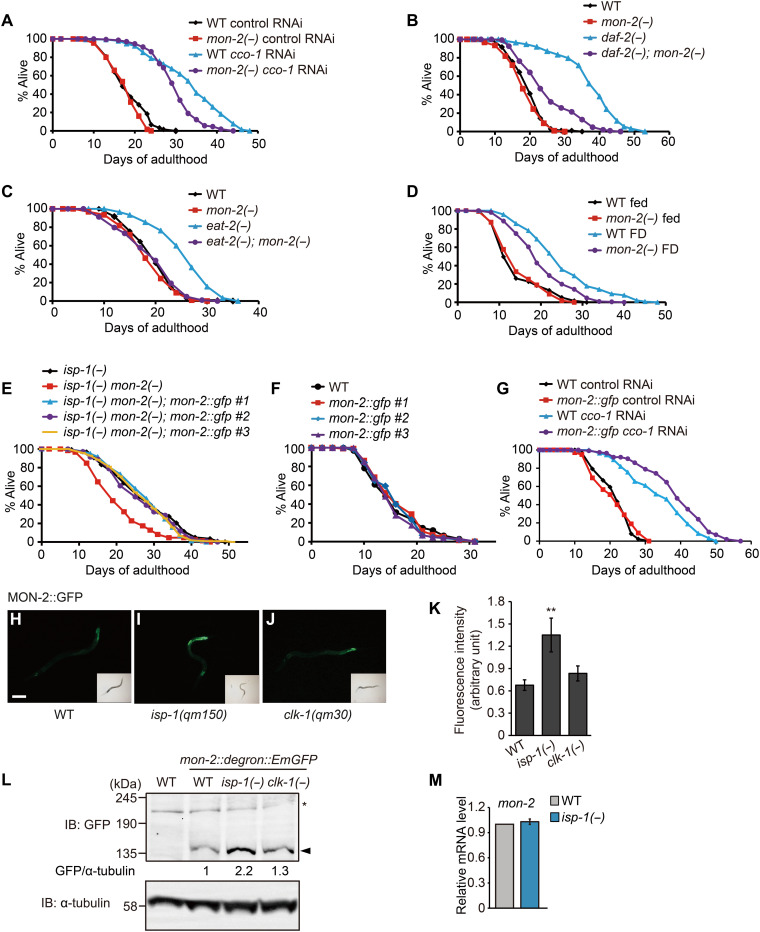
Fig. 3. MON-2 is required for longevity conferred by various interventions.
(A to D) mon-2(−) mutations reduced the long life span caused by cco-1 RNAi (A), daf-2(e1370) [daf-2(−)] (B), eat-2(ad1116) [eat-2(−)] (C), or food deprivation (FD) (D). (E and F) Transgenic expression of three independent lines of mon-2p::mon-2::gfp rescued the short life span of isp-1(qm150) mon-2(xh22) [isp-1(−) mon-2(−)] mutants (E), while not affecting the life span of WT animals (F). (G) Transgenic expression of mon-2p::mon-2::gfp line #1 further increased the long life span of cco-1 RNAi–treated animals. (H to J) Fluorescent signals of MON-2::GFP (green fluorescent protein) in WT (H), isp-1(qm150) [isp-1(−)] (I), and clk-1(qm30) [clk-1(−)] (J) animals. L2 or L3 larvae were used for imaging. Scale bar, 100 μm. (K) Quantification of (H to J) (N > 29 animals per condition). Error bars represent the SEM (**P < 0.01, two-tailed Student’s t test). (L) Western blot analysis of MON-2 protein using mon-2::degron::EmGFP CRISPR-mediated knock-in animals in WT, isp-1(−) or clk-1(−) mutant backgrounds (n = 3). α-Tubulin was used as a loading control [note that the MON-2::degron::EmGFP band was observed at around 135 kDa, which is unexpected, as the expected size is 212 kDa; because this study is the first one for detecting an endogenously tagged C. elegans MON-2, we speculate that the 135-kDa band was derived from either a posttranslationally modified (e.g., cleaved) MON-2 or a newly identified isoform of MON-2.]. Filled arrowhead indicates MON-2::degron::EmGFP, and the asterisk indicates a nonspecific band. IB, Immunoblot. (M) isp-1(−) did not affect mon-2 mRNA levels. Error bars represent SEMs (n = 3, two-tailed Student’s t test). See tables S3 and S4 for specific values and statistical analyses of the life-span data shown in this figure.