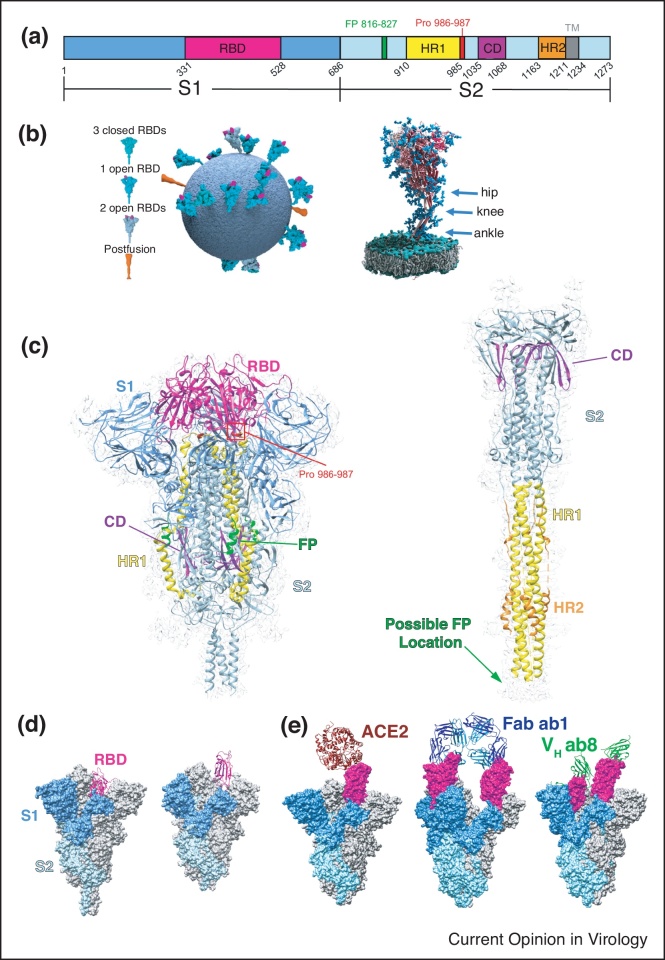
Figure 2.
Structures of SARS-CoV-2 spikes. (a) Schematic representation of the SARS-CoV-2 S domains showing S1 subunit(blue), S2 subunit (light blue), Receptor binding domain (RBD, magenta), fusion peptide (FP, green), heptad repeat 1 (HR1, yellow), connector domain (CD, purple), heptad repeat 2 (HR2, orange) and the transmembrane region (TM, grey). (b) Model of the SARS-CoV-2 virion showing the conformations and flexibility of S on the surface of virions. Reprinted by permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Nature, Ke et al. [23••]: Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Copyright 2020 [19]. Three flexible hinges are marked by arrows on the right. Adapted from Ref. [26•] under CC BY 4.0. (c) Single-particle cryoEM structure of the S trimer in its prefusion (PDB 6XR8 and EMD-22292) and postfusion (PDB 6XRA and EMD-22293) conformations with structural components colored to match the color scheme in (a) [32••]. (d) Comparison of S with RBD in the ‘down’ (left, PDB 6XR8) [32••] and ‘up’ (middle, 6VYB) [62] conformations. (e) CryoEM structures of S bound to ACE2 (left, PDB 7DF4) [34•] with RBD in the ‘up’ conformation, SN501Y mutant bound by Fab ab1 (middle, PDB 7MJJ) [33] with RBD in the ‘up’ conformation, and SN501Y bound by VH ab8 in both the RBD ‘up’ and ‘down’ conformations (7MJH) [33].