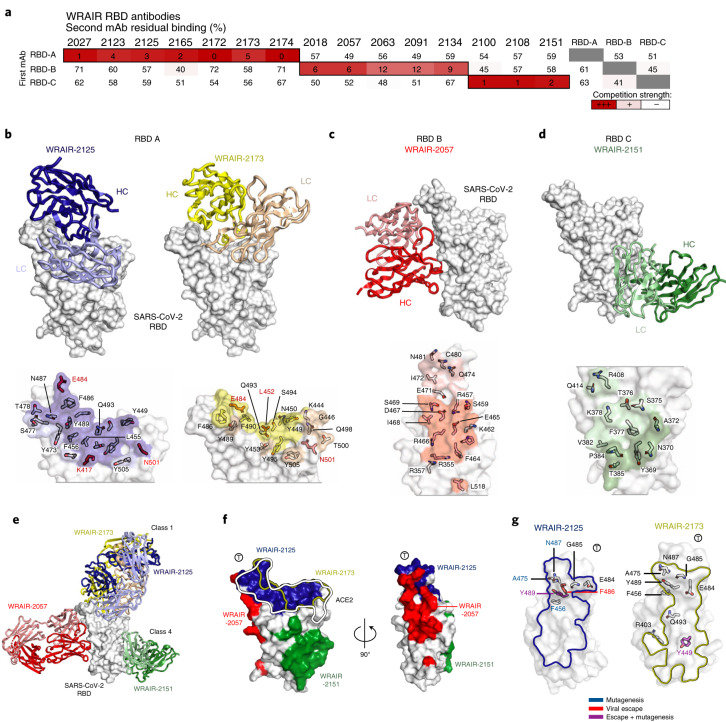
Fig. 3. Structure and epitope determination of SARS-CoV-2 RBD-targeting mAbs.
a, Epitope binning of RBD-directed mAbs via a BLI-based competition assay. Values represent the percentage of residual binding of the indicated second antibody after saturation of the antigen (RBD molecule) with the indicated first antibody. Shading from dark to light red indicates competition strength ranging from strong (0–25%), to intermediate (25–50%), to lack thereof (>50%). Competition groups are indicated by black boxes. Control antibodies RBD A, RBD B and RBD C were CC12.1, CC12.16 and CR3022, respectively. b–d, Top, representative crystal structures of RBD-targeting antibodies for WRAIR RBD groups A, B and C are shown. RBD A mAbs, WRAIR-2125 (dark blue) and WRAIR-2173 (yellow) target the ACE2 binding site. RBD B mAb, WRAIR-2057 (red) recognizes a novel epitope on the ‘side’ of the RBD distal from the ACE2 binding site centered on residue E465. RBD C mAb, WRAIR-2151 (dark green) targets a CR3022-like site on the RBD. Bottom, epitope footprints of respective antibodies are shown on the surface of the RBD and colored based on the antibody heavy-chain and light-chain colors. RBD contacting residues are shown as sticks, with residues seen in VOCs highlighted in bold red. e, Structures of WRAIR RBD A, B and C antibodies are shown on a single RBD molecule to highlight the different recognition modes. f, RBD A, B and C epitopes are shown on the RBD surface with the ACE2 binding interface highlighted by the black/white line. g, Epitope mapping of WRAIR-2125 and WRAIR-2173 contact residues identified in the shotgun mutagenesis (blue) and viral escape experiments (red), or both (purple) are shown in stick representation.