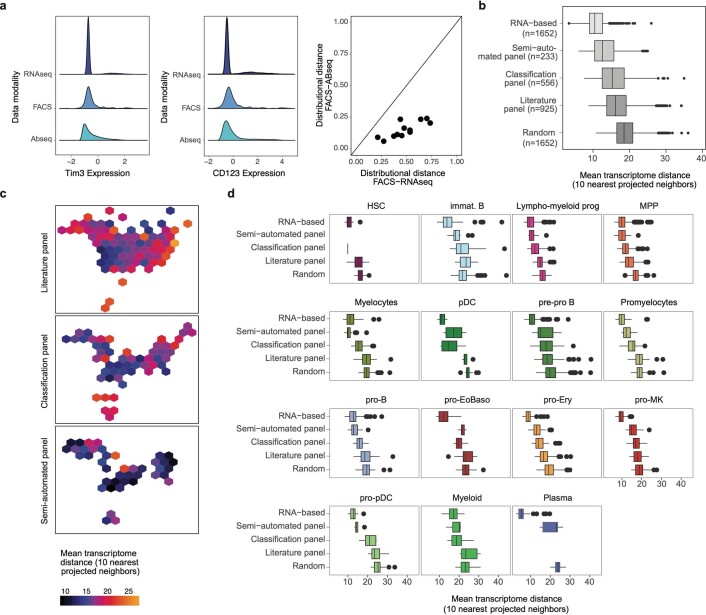
Extended Data Fig. 10. Projection and classification of cytometry data using a single-cell proteo-genomic reference.
Related to Fig. 7. a. Distribution of normalized, scaled expression values of Tim3 (left panel) and CD123 (central panel) measured by scRNA-seq, Abseq, and FACS. Right panel: Scatter plot depicts the dissimilarity between the distribution of expression values measured by FACS, and the distribution measured by scRNA-seq (x-axis) or Abseq (y-axis) as quantified using Kolmogorov-Smirnov distance. Data for all markers included in the panel from main Fig. 6f is shown. b–d. Comparison of data integration strategies. Smart-seq2 data and Abseq data were integrated with five different strategies. RNA-based: Integration by Seurat v3, based on gene expression (transcriptome). Random: Random selection of ten nearest neighbors. Others: Surface marker-based integration using NRN, using defined sets of surface markers (Classification panel, Semi-automated panel: see Supplementary Table 6. Literature panel: CD34, CD38, CD45RA, CD90, CD10, CD135/FLT3, CD49f). For every cell projected on the UMAP, the ten nearest neighbors in projected UMAP space were identified. Subsequently, the mean Euclidean distance between their location in a gene expression-based PCA space (Smart-seq2) was computed. Sample size n = 1652. b. Boxplot summarizing the distance across data integration strategies. See figure for sample size. See Methods, section ‘Data visualization for a definition of boxplot elements’. c. Hexagonal plot summarizing the projection accuracy for different regions of the UMAP. d. Boxplots stratified by cell type demonstrate that projection using the semiautomated panel performs close to an RNA-based integration in most cases. See panel b for sample size.