Figure 3. Structural and sequence comparison of AMPA receptor auxiliary subunits.
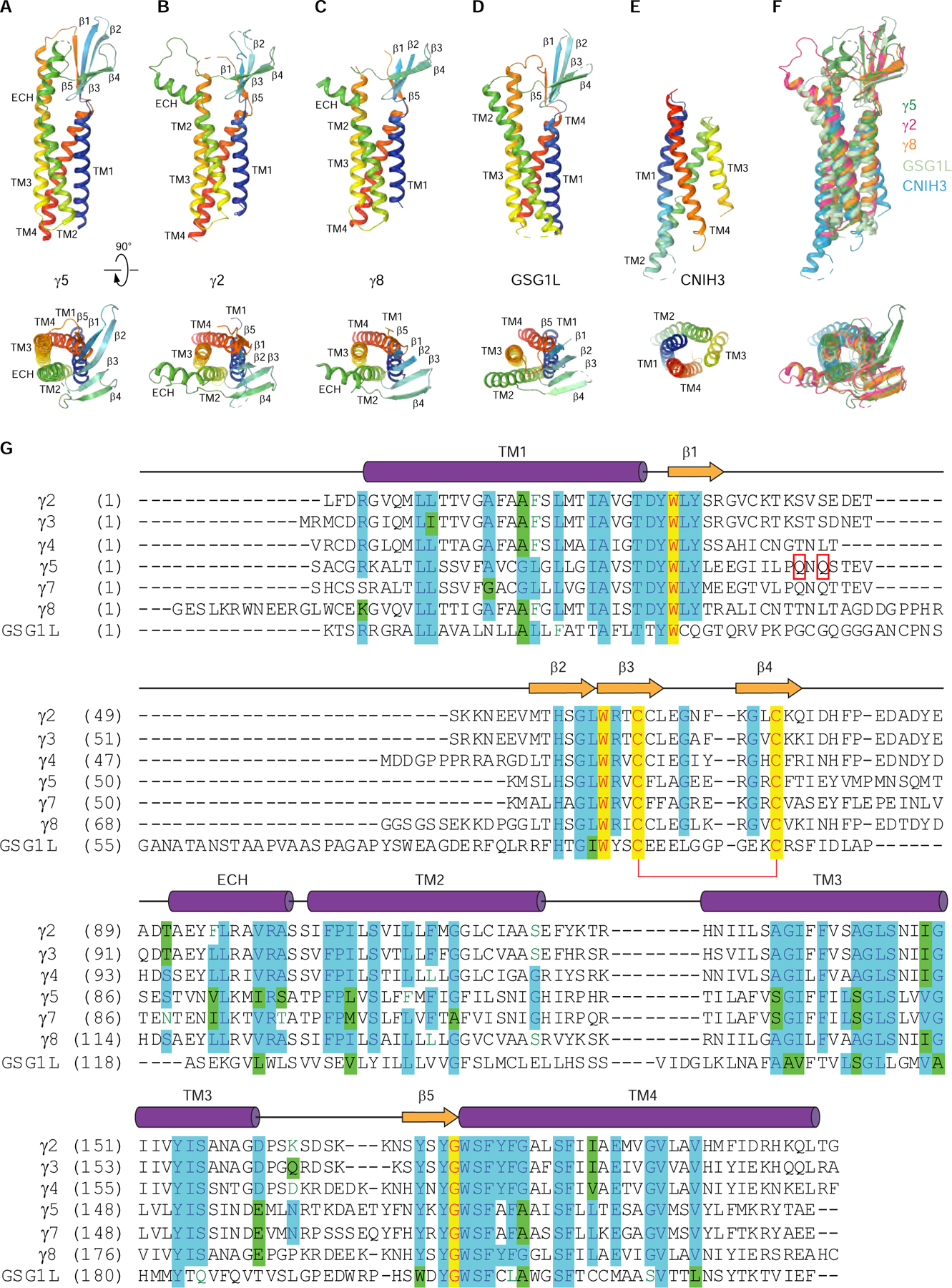
(A-E) Rainbow-colored (from blue N-terminus to red C-terminus) structures of auxiliary subunits γ5 (A, this study), γ2 (B, PDB ID: 5WEO), γ8 (C, PDB ID: 6QKC), GSG1L (D, PDB ID: 5WEL) and CNIH3 (E, PDB ID: 6PEQ) viewed parallel to the membrane (top row) or extracellularly (bottom row).
(F) Superposition of γ5 (blue), γ2 (red), γ8 (orange), GSG1L (cyan) and CNIH3 (green).
(G) Amino acid sequence alignment of AMPA receptor auxiliary subunits γ2 (Uniprot ID Q88602), γ3 (Q9JJV5), γ4 (Q9JJV4), γ5 (Q8VHW4), γ7 (P62956), γ8 (Q8VHW5) and GSG1L (D3Z7H4). The secondary structure is shown for γ2 above the sequence alignment as cylinders (α-helices), arrows (β-strands) or lines (loops). Completely conserved residues are highlighted in yellow. Mostly conserved residues are highlighted in blue (or green for homologous residues). Conserved cysteines forming a disulfide bridge between β3 and β4 are connected by a red bracket. Residues in the β1-β2 loop that are in close proximity to residues K695 and K697 in GluA2 LBD are indicated by red rectangles. The C-terminal residues are excluded.
