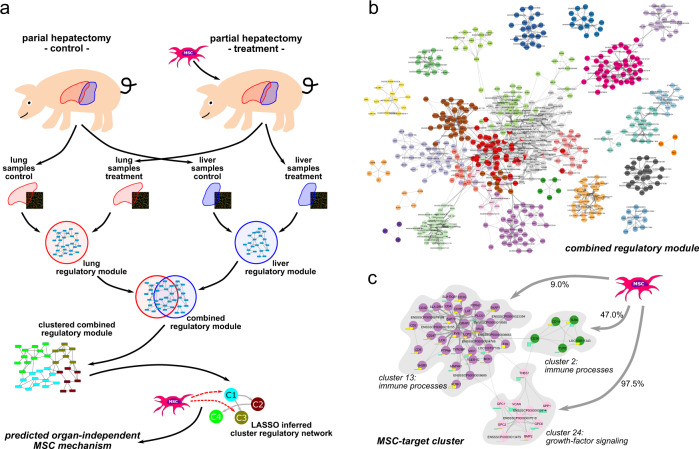
Fig. 2. Schematic workflow for the prediction of organ-independent molecular MSC targets.
a Gene expression was measured in lung and liver samples after partial hepatectomy of control and MSC-treated pigs. ModuleDiscoverer was used to identify organ-specific regulatory modules of the response to MSC treatment. Topological clustering of the combined regulatory module was performed to identify clusters of proteins with similar biological functions. Based on the average cluster expression profiles for lung and liver samples, and MSC treatment as additional perturbation, we computed a cluster regulatory network. The predicted relations between MSC and the clusters resemble potential mechanisms of MSC action. b Combined regulatory module composed of the union of the lung-specific and the liver-specific regulatory modules. For reading of individual protein members in the 29 clusters (please cf. Supplementary Data 1). c The STRING-network outlining all proteins that belong to clusters predicted as targeted by MSC. Cluster membership of proteins is color-coded. For each protein, log2-FC for the liver (left) and lung (right) of the associated gene is shown as chart next to the protein. Up-/downregulation is colored yellow/turquoise, respectively.