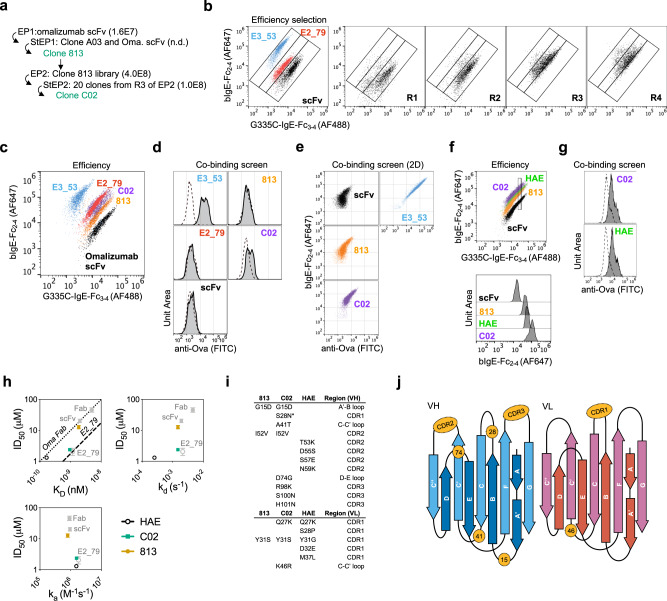
Fig. 3. Omalizumab variants exhibit enhanced affinity and disruptive efficiency.
a Overview of selections with number of transformants per library in brackets. Full details of selections outlined in Supplementary Fig. 2. b Controls and freshly induced R1–4 of EP1 library stained with disruption efficiency stain. c Hits and controls were stained with disruption efficiency stain, followed by fixation. d Histogram of yeast hits and controls stained for co-binding:100 nM bIgE-Fc2–4 (dotted line) or precomplexed bIgE-Fc2–4:FcεRIα-Ova (100 nM:1 μM-solid line), with fixation. e Two color co-binding screen of omalizumab variants stained with bIgE-Fc2–4:FcεRIα-Ova (100 nM:1 μM) to assess correlation between bIgE-Fc2–4 and FcεRIα-Ova signals as compared to a non-competitive control (E3_53). f HAE yeast stained with disruption efficiency stain with fixation compared to omalizumab variants. For clarity a fraction of G335C-IgE-Fc3–4 positive cells were gated by their G335C-IgE-Fc3–4 staining intensity, and the relative intensity of biotin-IgE-Fc2–4 was displayed by histogram. g C02 and HAE stained with co-binding stain with fixation. h ID50 with 95% CI from fits in Supplementary Fig. 3d vs. KD, ka, or kd by variant. The efficiency ratio for omalizumab Fab and E2_79 are plotted as benchmarks for other variants (dotted and dashed line). i Amino acid mutations in clones 813, C02, and HAE relative to omalizumab. j Distribution of mutations in (i) mapped onto topology of VH and VL domains.