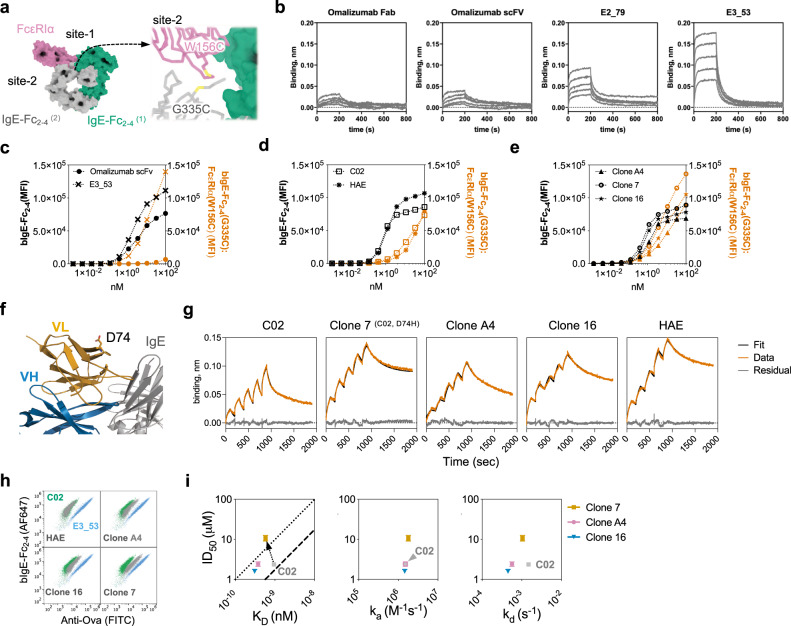
Fig. 5. A disulfide “locked” IgE-Fc2–4:FcεRIα complex enables the selection of omalizumab variants that trap a disruption intermediate.
a IgE-Fc2–4:FcεRIα complex (2Y7Q), with the interface of FcεRIα and IgE at “site-2” revealed to display position of IgE-Fc2–4 (G335C) and FcεRIα (W156C) mutations. b Octet binding studies with locked-complex on tips exposed to anti-IgE agents as indicated in a twofold serial dilution from 400 nM to 25 nM. c Binding profiles of yeast-displayed omalizumab scFv and E3_53 to free bIgE-Fc2–4 (left axis, black) or locked-complex (right axis, red). d Same as in c for yeast-displayed C02 and HAE. e Same as in c for clones A4, 7, 16. f Schematic of novel E-stand mutation position relative to omalizumab:IgE complex structure. g BLI SCK binding studies of anti-IgE variants as indicated with locked-complex on tips with anti-IgE agents in a twofold serial dilution from 1600 nM to 100 nM. h Yeast-displayed omalizumab clones 7, A4, and 16 stained with bIgE-Fc2–4 (100 nM):FcεRIα-Ova (1 μM) complexes relative to yeast expressing C02, HAE, and E3_53 (samples fixed). i ID50 with 95% CI from fits in Supplementary Fig. 5h vs. KD, ka, kd for A4, 7, and 16. The efficiency ratio for omalizumab Fab and E2_79 are plotted as benchmarks for variants (dotted and dashed line). Source data are provided as a Source Data file.