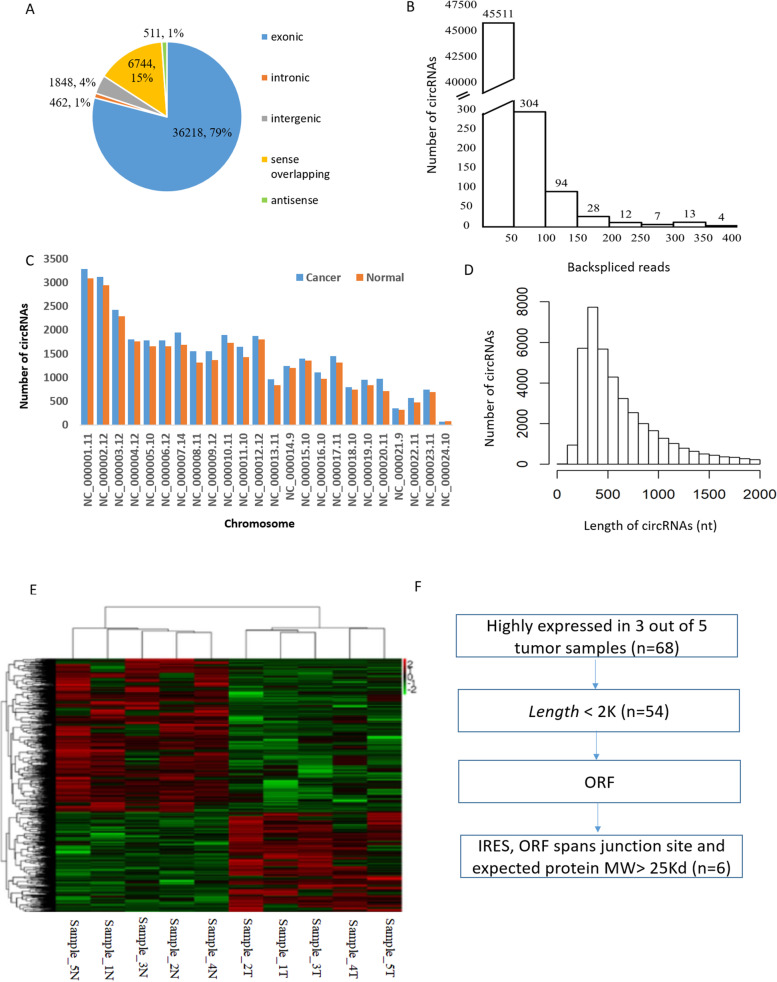
Fig. 1.
Differential expression profile of circRNAs in human gastric cancer (GC) and adjacent normal tissues. a The numbers and ratios of circRNAs originating from different regions of the genome. b The number of circRNAs and back-spliced reads identified. X-axis: the backspliced reads of circRNAs detected in this study. Y-axis: the number of circRNAs. c The chromosome distribution pattern of circRNAs shows no difference between the cancer and normal groups. d The length of the majority of the circRNAs is less than 1500 nucleotides (nt). e The dendrogram shows the relationship between the samples and the differentially expressed circRNAs. The differential expression in the normal and cancer samples was clearly distinguishable. f The workflow used to identify functional circRNAs with coding potential in GC