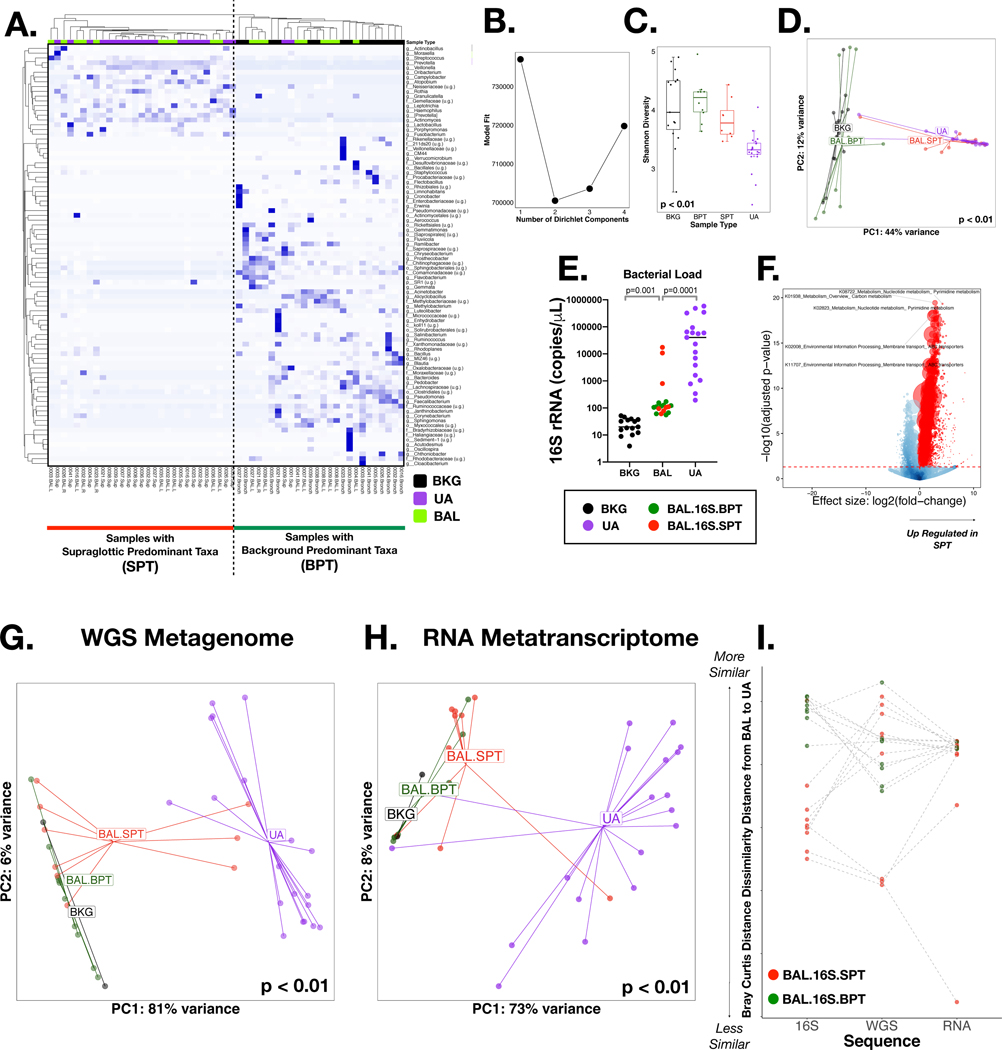
Figure 1: 16S rRNA gene, Whole genome (WGS) and RNA sequencing:
Background (BKG), Upper Airway (UA) and Bronchoalveolar (BAL) samples were collected via bronchoscopy; 16S rRNA gene, Whole genome and RNA sequencing was performed. (A) A heatmap based on Bray-Curtis distance for the 16S rRNA gene sequencing, illustrates the top taxa for all samples. Hierarchical clustering showed two clear clusters, one with BKG samples and BAL samples similar to BKG (Background Predominant Taxa) and another with UA samples and BAL samples similar to UA (Supraglottic Predominant Taxa). (B) Dirichlet Multinomial Modelling (DMM) showed 2 clusters had the best model fit for the 16S rRNA gene sequencing. (C) α Diversity, measured by Shannon Index, showed significant (Wilcoxon) difference between all samples and lowest diversity in UA and among BAL samples that clustered to BAL.16S.SPT by DMM. (D) Beta Diversity, measured by Bray-Curtis, also indicates a significant (PERMANOVA) difference between all samples for 16S rRNA gene sequencing. (E) Bacterial load, measured by ddPCR showed highest levels in UA samples (Kruskal-Wallis). BAL Samples also had higher levels when compared to BKG samples. (F) The inferred metagenome was assessed using PICRUST highlighting several significantly enriched pathways (colored in red). (G) β Diversity for WGS, measured by Bray-Curtis, showed a significant (PERMANOVA) difference between all samples, with UA samples separate from BKG and BAL.BPT samples. Three BAL.SPT samples clustered with UA Samples. (H) β Diversity for RNA, measured by Bray-Curtis, showed a significant (PERMANOVA) difference between all sample types. Two BAL.SPT samples clustered with UA samples. (I) Z Transformed Bray-Curtis Distance between BAL samples and paired UA samples showed clear separation of BAL.16S.BPT and BAL.16S.SPT samples in 16S rRNA gene sequencing. This separation was not as clear in WGS and RNA.