Abstract
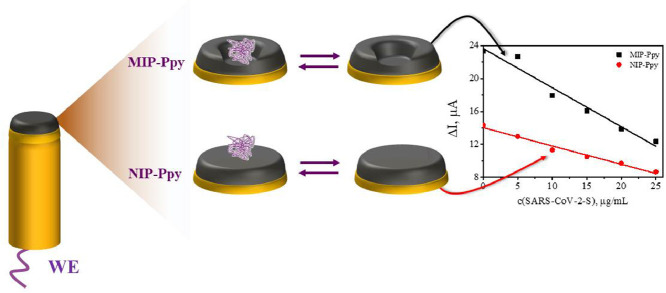
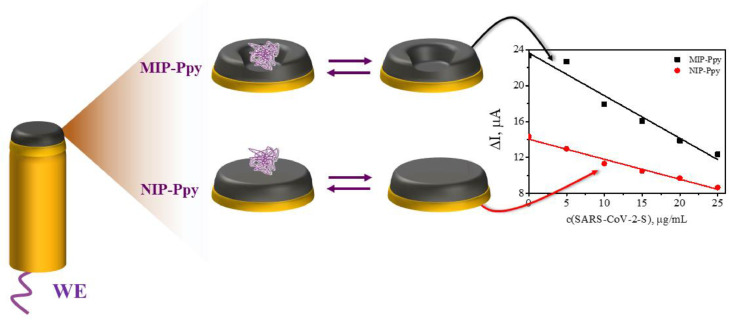
This study describes the application of a polypyrrole-based sensor for the determination of SARS-CoV-2-S spike glycoprotein. The SARS-CoV-2-S spike glycoprotein is a spike protein of the coronavirus SARS-CoV-2 that recently caused the worldwide spread of COVID-19 disease. This study is dedicated to the development of an electrochemical determination method based on the application of molecularly imprinted polymer technology. The electrochemical sensor was designed by molecular imprinting of polypyrrole (Ppy) with SARS-CoV-2-S spike glycoprotein (MIP-Ppy). The electrochemical sensors with MIP-Ppy and with polypyrrole without imprints (NIP-Ppy) layers were electrochemically deposited on a platinum electrode surface by a sequence of potential pulses. The performance of polymer layers was evaluated by pulsed amperometric detection. According to the obtained results, a sensor based on MIP-Ppy is more sensitive to the SARS-CoV-2-S spike glycoprotein than a sensor based on NIP-Ppy. Also, the results demonstrate that the MIP-Ppy layer is more selectively interacting with SARS-CoV-2-S glycoprotein than with bovine serum albumin. This proves that molecularly imprinted MIP-Ppy-based sensors can be applied for the detection of SARS-CoV-2 virus proteins.
Keywords: COVID-19, SARS-CoV-2 spike glycoprotein, Polypyrrole (Ppy), Conducting polymers, Molecularly imprinted polymers (MIPs), Electrochemical determination of virus proteins, Thin layers
Graphical abstract
1. Introduction
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) induced COVID-19 pandemic that began in 2019 has caused drastic changes in the world. 197 countries were affected [1]: lockdowns [2], quarantine, economic problems hit the most significant part of the world, people's emotional health has deteriorated. Even at the beginning of the 2021, this pandemic is still not adequately controlled. Although the vaccines became available to society, this viral infection is still very active and the virus is rather rapidly mutating and appears in new even more infectious forms. Therefore, a much deeper understanding of the virus SARS-CoV-2 is required and rapid analytical methods that are suitable for the diagnosis of COVID-19 and/or detection of virus or their parts are demanded to overcome and defeat this infection. Thus, various aspects of the virus itself [3], genome [4], [5], [6], [7], research of the structure, function of proteins, and nucleocapsid, envelope, spike, and membrane protein interactions with drugs [8], [9], [10], [11], and some other aspects [12, 13] were investigated. Better and easier detection methods could improve the diagnosis of viral infection and enable more efficient ways of defeating the COVID-19 pandemic. Recently, label-free protein detection has become relevant in research and clinical practice [14, 15]. The discovery and detection of biomarkers during the diagnosis of human diseases is required for biomedical purposes [15, 16].
In biosensors, the analyte recognition elements are typically based on bio-macromolecules such as enzymes, antibodies, DNA, aptamers, etc. However, such bioanalytical systems have some limitations due to operating conditions and expensive production. Therefore, the development of artificial biorecognition-systems based on synthetic receptors and molecularly imprinted polymers (MIPs) has attracted a great interest as a potential alternative [14, [17], [18], [19]]. Researchers have been focused on the development of a system that replicates the natural recognition process. Therefore, the interest in the development of MIPs has grown during recent years [15, 16, [20], [21], [22], [23], [24], [25], [26], [27]]. The technique of molecular imprinting allows the formation of specific molecular recognition sites that operate on the principle of complementarity between the imprinted sites and the analyte. Therefore, MIPs can selectively bind the analytes of interest, which were used as templates during formation of these MIPs [14, 16, [28], [29], [30]]. MIPs also have some benefits including low-cost, easy way of preparation, advanced storage stability, and rather good specificity [14, 31]. In previous studies, it was reported that various types of small molecules can be imprinted within polymers [24, 29, 32, 33]. In some researches, it was demonstrated that high molecular mass biomolecules including proteins [15, 22, 23, [34], [35], [36], [37], [38], [39], [40], [41], [42], [43]] can be also molecularly imprinted within polymers. Polypyrrole (Ppy) is among several other polymers that can be very efficiently applied for the design of MIP-based sensors [24, 29, 32, 33, [44], [45], [46], [47]]. Ppy is a conducting polymer, which can be easily electropolymerized and used as a polymeric matrix of MIPs for the detection of low and high molecular weight analytes [15, 44]. Electrochemical methods like cyclic voltammetry, differential pulse voltammetry, and electrochemical impedance spectroscopy were used for the detection of the proteins both on the polypyrrole modified with molecular imprints and on the unmodified in previous studies [15, [48], [49], [50], [51], [52], [53], [54]]. Meanwhile, there is only few reports on the application of chronoamperometry for determination of virus-proteins [44]. In chronoamperometry the changes in the current appear in response to increase or decrease of the diffuse layer thickness at the surface of the working electrode [55]. Therefore, the application of chronoamperometry (in pulsed amperometric mode) and the analysis of data gathered by this method using Cottrell or Anson plots are providing interesting and useful insights into the evaluation of interaction between analytes and the electrode [56].
At the moment, there are some explorations reported that are already applying MIP technology for SARS-CoV-2 [57, 58]. The development of so called ‘monoclonal-type plastic antibodies’ based on MIPs was described [57]. Such ‘antibodies’ were able to selectively bind a spike protein of the novel coronavirus SARS-CoV-2 to block its function. The obtained nanoparticles were analyzed by SDS-PAGE electrophoresis. The results of the electrophoretic analysis demonstrated promising results in the formulation of ‘free-drug therapeutics’ due to their ability to bind the virus spike glycoprotein and, thus, to block the infection process. According to reported results it was concluded that the ‘monoclonal-type plastic antibodies’ could be potentially used as free-drug therapeutics in the treatment of infection by novel coronavirus (2019-nCoV). In another research, SARS-CoV-2 nucleoprotein (ncovNP) was qualitatively and quantitatively determined by MIP-based layer on poly-m-phenylenediamine (PmPD), which was deposited on the Au-TFE electrode [58]. Cyclic voltammetry (CV) was applied for the characterization of the preparation steps of the sensor. Meanwhile, the rebinding of SARS-CoV-2 nucleoprotein on the sensors was studied by differential pulse voltammetry (DPV) in the solution of 1 M KCl containing a redox probe K3[Fe(CN)6]/K4[Fe(CN)6]. The obtained results demonstrated the linear increase of the sensor response with increasing ncovNP concentration. The feasibility of sensor performance in clinical samples was tested. For this purpose, they analyzed the samples prepared from nasopharyngeal swab specimens. Genetically engineered receptor-binding domain of SARS-CoV-2-RBD protein was imprinted in ortho-phenylenediamine and deposited on a macroporous gold screen-printed electrode [59].
The aim of our recent research was to design the MIP-based sensor for the determination of SARS-CoV-2-S glycoprotein. For this purpose, Ppy layers were deposited on the working platinum electrode from the polymerization mixture containing SARS-CoV-2-S glycoprotein and pyrrole dissolved in phosphate buffered saline (PBS) solution, pH 7.4. The performance of the electrode modified by the deposited MIP-Ppy layer imprinted with SARS-CoV-2-S glycoprotein was investigated and compared with that of non-imprinted (NIP-Ppy) layer.
2. Materials and methods
2.1. Chemicals and instrumentation
Pyrrole 98% (Alfa Aeser, Germany), H2SO4 (96%) (Lachner, Czech Republic), HNO3, NaOH (Merck, Germany), H2PtCl6 (Merck, Germany), and bovine serum albumin (BSA) (Carl Roth, Germany) were used as received. KH2PO4 (Honeywell Riedel-de Haen, Germany), NaCl, KCl, and Na2HPO4 (Roth, Germany) salts were used for the preparation of buffer. The detailed description of expression and purification of SARS-CoV-2-S spike glycoprotein is presented in supporting material.
Experiment was performed using potentiostat/galvanostat Metrohm AutoLAB model µAutolabIII/FRA2 µ3AUT71079 controlled by NOVA 2.1.3 software (EcoChemie, The Netherlands). All measurements were done in a homemade cell. The total volume of the cell was 250 µL. Three-electrode system consisted of Pt disk with 1 mm diameter sealed in glass as the working electrode, Ag/AgCl in 3 M KCl solution electrode as a reference electrode (Ag/AgCl), and Pt disk of 2 mm diameter as a counter electrode.
2.2. Pretreatment of working electrode
The working electrode was pretreated before electrochemical deposition of Ppy following the procedure described in previous studies [44, 60]. All solutions were thoroughly degassed just before use with a stream of nitrogen (N2). According to this procedure, the Pt electrode was rinsed with concentrated HNO3 solution in an ultrasonic bath for 10 min, then rinsed with water and polished with alumina paste. Later, it was rinsed with water again and then with 10 M solution of NaOH, thereafter – with 5 M solution of H2SO4 in an ultrasonic bath for 5 min. Electrochemical cleaning of the electrode was carried out in 0.5 M H2SO4 by cycling the potential for 20 times in the range between −100 mV and +1200 mV vs Ag/AgCl at a sweep rate of 100 mV s − 1. The identification of the bare electrode surface was made possible by a stable indication of the cyclic voltammogram. To improve the adhesion of the Ppy layer to the electrode surface, a layer of ‘platinum black’ was deposited over the working electrode [60]. Deposition of Pt clusters was performed in 5 mM solution of H2PtCl6 containing 0.1 M of KCl by 10 potential cycles in the range between +500 mV and −400 mV vs Ag/AgCl at a sweep rate of 10 mV s − 1.
2.3. The electrochemical deposition of MIP and NIP and evaluation of sensor signal
The electrochemical deposition of the polypyrrole layer was performed in the same electrochemical cell. NIP-Ppy was electrochemically deposited from the polymerization solution containing 0.5 M solution of pyrrole in PBS. The preparation of MIP-Ppy was carried out in two steps. Step I: deposition of polymeric layer was carried out from the polymerization solution containing 0.5 M solution of pyrrole and 50 μg/mL of SARS-CoV-2-S glycoprotein all dissolved in PBS solution. The polymeric layers were formed by a sequence of 20 potential pulses of +950 mV for 1 s, between these pulses 0 V potential for 10 s was applied [44, 60]. Step II: the MIP-Ppy was formed when the imprinted protein molecules were extracted by incubation in 0.05 M H2SO4 for 10 min. In the same way as MIP-Ppy, NIP-Ppy was also exposed to 0.05 M solution of H2SO4. MIP-Ppy and NIP-Ppy were analyzed using pulsed amperometric detection by the sequence of 10 potential pulses of +600 mV vs Ag/AgCl lasting for 2 s, between these pulses 0 V vs Ag/AgCl was applied for 2 s (Fig. 1 ).
Fig. 1.
Schematic representation of evaluation by chronoamperometry of Pt electrode modified with non-imprinted polypyrrole (NIP-Ppy) and with molecularly imprinted polypyrrole (MIP-Ppy) with SARS-CoV-2-S glycoprotein imprints. Electrochemical measurements were performed in phosphate-buffered saline (PBS) solution, pH 7.4.
3. Results and discussions
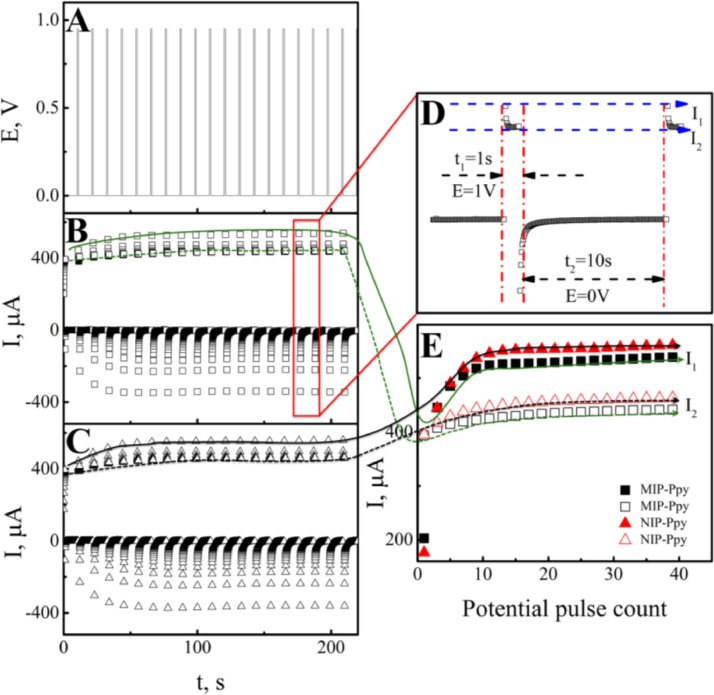
Electrochemical polymerization of the two types of Ppy layers was performed by a sequence of potential pulses (Fig. 2 ). The profile of potential pulses sequence is represented in Fig. 2A. Figs. 2B and 2C demonstrate the currents registered during the electrochemical deposition of Ppy layer from polymerization solution containing SARS-CoV-2-S glycoprotein and Ppy layer from polymerization solution without SARS-CoV-2-S glycoprotein on Pt-electrode surface.
Fig. 2.
Electrochemical deposition of the polypyrrole layers on the Pt electrode: A – The profile of potential applied during the sequence of potential pulses; B – The profile of current registered during the deposition of Ppy layer from polymerization solution containing SARS-CoV-2-S glycoprotein; C – The profile of current registered during the formation of Ppy layer from polymerization solution without SARS-CoV-2-S glycoprotein. D – The profile of current registered during one potential pulse. E – Changes of current measured instantly after a potential step of +950 mV.
The changes of current at the beginning I1 and at the end I2 of pulses of the potential at +950 mV are presented in Fig. 2E. The current changes at the potential of 0 V were not the object of analysis, because during this potential step the equilibration of monomer and template molecule concentrations in the neighborhood of the working electrode is happening. Previous studies demonstrated that the self-assembly of monomers and template molecules due to the interactions under thermodynamic control prior to polymerization, is significant for the recognition characteristics of the final polymers [61]. Polymerization of Ppy occurs during the pulses at a potential value of +950 mV. Therefore, only an insignificant Faradaic process was observed on the electrode at the 0 V potential step. Thus, the current changes during the potential step when the potential was elevated up to +950 mV were analyzed in more detail. For the visualization of the current changes during the electrochemical deposition of Ppy layer from polymerization solution containing SARS-CoV-2-S glycoprotein and Ppy layer from polymerization solution without SARS-CoV-2-S glycoprotein, two current points at the beginning I1 and end I2 of each potential step were taken into account (Fig. 2D). The comparison of the current changes demonstrated that the current registered during deposition of Ppy layer from polymerization solution without SARS-CoV-2-S glycoprotein is higher than that registered during deposition of Ppy layer from polymerization solution containing SARS-CoV-2-S glycoprotein (Fig. 2E). However, the observed difference of current changes is not very significant in comparison with that registered in our previous researches [29] and in other researches [29, 62]. The collation of current changes on Pt electrode during the electrochemical deposition of Ppy/SARS-CoV-2-S and NIP-Ppy layers have illustrated that current during the deposition of NIP-Ppy increased just by 1.05 times in comparison to that registered during the deposition of Ppy/SARS-CoV-2-S. From the current changes observed during the polymerization, it can be presumed that the entrapped protein molecules just insignificantly affect the conductivity of the formed layers. During the next MIP-Ppy preparation step, the entrapped SARS-CoV-2-S glycoproteins were removed from the formed Ppy/SARS-CoV-2-S layer and MIP-Ppy was formed. In the same way, as MIP-Ppy, NIP-Ppy was also exposed to 0.05 M H2SO4 to eliminate any differences caused by the extraction procedure on the formed MIP-Ppy, NIP-Ppy layer properties.
In the following part of the research, the formed MIP-Ppy and NIP-Ppy layers were evaluated using pulsed amperometric detection by a sequence of 10 potential pulses of +600 mV and 0 V for 2 s each as it was suggested in our previous research [44]. Various aspects of charging-discharging of conducting polymer polypyrrole were well discussed by Heinze et al. [63]. Also, there was stated that overoxidation of the un-substituted Ppy already occurs at 0.65 V vs Ag/AgCl(3 M KCl) [64]. Hence, taking into account these findings potential pulse values of 0 V and +600 mV were selected for the determination of SARS-CoV-2-S glycoproteins.
The profile of the potential pulse sequence is presented in Fig. 3 A.
Fig. 3.
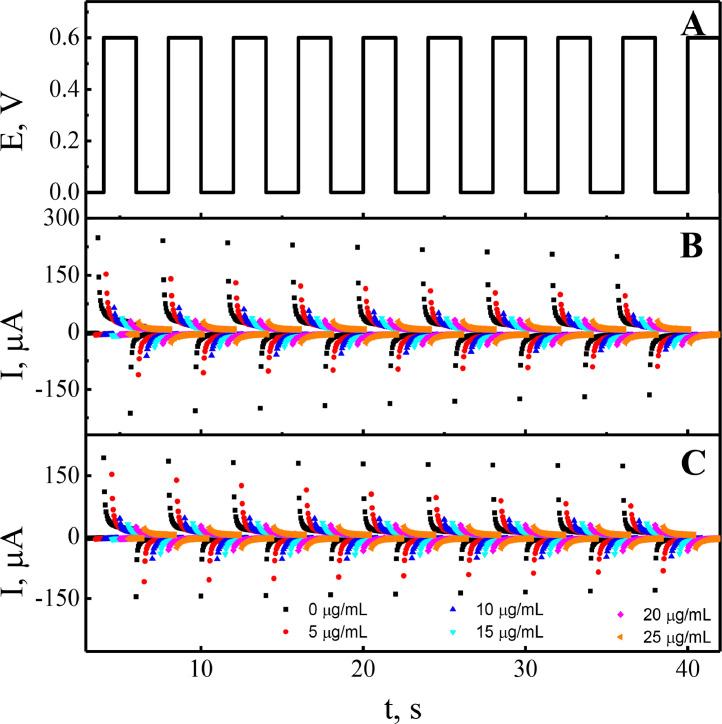
Electrochemical evaluation of MIP-Ppy and NIP-Ppy layers was performed by the potential pulse sequence. A – potential pulse profile. Typical chronoamperograms (during pulsed amperometric detection) were obtained at: B – MIP-Ppy and C – NIP-Ppy modified Pt electrodes in the absence of SARS-CoV-2-S glycoprotein (•) and in the presence of SARS-CoV-2-S glycoprotein from 5 μg/mL up to 25 μg/mL in PBS solution, pH 7.4 (offset 0.5).
The concentration of SARS-CoV-2-S glycoprotein was varying in the range from 0 μg/mL to 25 μg/mL. Some other reports described instability of the proteins in presence of salts [65, 66], but during the preparation of required concentrations no signs of instability of the SARS-CoV-2-S glycoprotein solubilized in PBS were observed. Figs. 3B and C demonstrate the dependence of the chronoamperometric response (during pulsed amperometric detection) of MIP-Ppy and NIP-Ppy modified electrodes to SARS-CoV-2-S glycoprotein. The change in the chronoamperometric response is related to the adsorption of less conductive protein molecules on the MIP-Ppy and NIP-Ppy layers. When SARS-CoV-2-S glycoprotein concentration in solution was increased, the registered chronoamperometric response of both MIP-Ppy and NIP-Ppy-modified Pt electrodes decreased. Higher currents were registered before the incubation of electrode in SARS-CoV-2-S glycoprotein containing solution. This effect is determined by the presence of water molecules and electrolyte ions in the places where molecular imprints were formed. After the incubation in SARS-CoV-2-S glycoprotein containing solution, the ions of solvent and the electrolyte were replaced by the molecules of SARS-CoV-2-S glycoprotein and thus the registered current at the potential of +600 mV decreased.
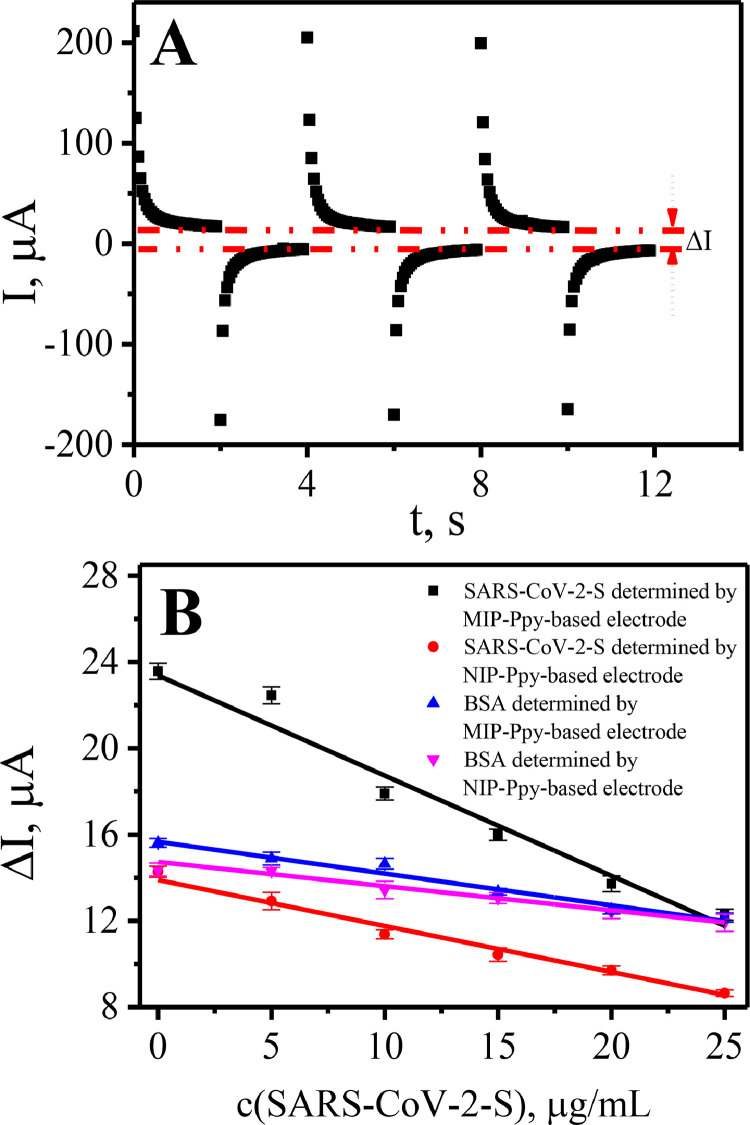
The magnitude of current differences, which are registered during potential pulses at instants when potentials were stepped from 0 mV up to +600 mV and +600 mV down to 0 mV, has decreased with increasing SARS-CoV-2-S glycoprotein concentration in PBS solution (Fig. 4 ). Fig. 4A represents the current profile, which was registered during potential pulses, and the way in which the analytical signals (ΔI) for the calibration curve was depicted. According to this calibration curve, linearity of analytical signal dependence on analyte concentration was observed at all evaluated SARS-CoV-2-S glycoprotein concentrations in the range from 0 μg/mL to 25 μg/mL.
Fig. 4.
Calibration curves of ΔI vs concentration of SARS-CoV-2-S glycoprotein and BSA on MIP-Ppy and NIP-Ppy according to the ΔI calculated in respect to: A – the principal of ΔI measuring; B – ΔI. RSD% was in range from 2 to 4.3% of current values of 5 potential pulses for the listed data points.
The slope derived using the linear regression equation for the changes of current (ΔI, μA) vs concentration of SARS-CoV-2-S glycoprotein (concentration expressed in μg/mL) registered by NIP-Ppy-modified Pt electrode was of −0.22 μA/(μg/mL) with R2 = 0.98 (Table 1 ). While the slope of linear regression for the Pt electrode modified with SARS-CoV-2-S glycoprotein imprinted MIP-Ppy was −0.47 μA/(μg/mL) with R2 = 0.96 (Table 1). The sensitivity calculated from the calibration curves of the MIP-Ppy modified Pt electrode towards SARS-CoV-2-S glycoprotein in the linear dependence interval according to the ΔI measurements was approximately 2.1 times higher than that of NIP-Ppy modified Pt electrode. This difference is significant and therefore can be applied in the design of sensors based on MIP-Ppy modified Pt electrodes.
Table 1.
Linear regression characteristics of current (ΔI, μA) vs concentration of SARS-CoV-2-S glycoprotein (c, μg/mL) on the MIP-Ppy and NIP-Ppy modified Pt electrodes.
| y = axe+b | a | b | R2 |
|---|---|---|---|
| SARS-CoV-2-S determined by MIP-Ppy-based electrode | −0.46±0.04 | 23.4 ± 0.7 | 0.96 |
| SARS-CoV-2-S determined by NIP-Ppy-based electrode | −0.21±0.01 | 13.9 ± 0.3 | 0.98 |
| BSA determined by MIP-Ppy-based electrode | −0.15±0.01 | 15.7 ± 0.2 | 0.97 |
| BSA determined by NIP-Ppy-based electrode | −0.10±0.01 | 14.7 ± 0.1 | 0.97 |
The same MIP-Ppy and NIP-Ppy modified Pt electrodes were evaluated for the interaction with BSA (Fig. 4B) to evaluate the selectivity of MIP-Ppy layer towards different proteins. The slope values for these measurements were derived using linear regression and they are represented in Table 1. The slope value (−0.15 μA/(μg/mL)) registered by the MIP-Ppy modified Pt electrodes incubated in BSA containing solution was significantly lower.
The comparison of the sensitivity/selectivity results among studies, which are reporting MIPs sensors based on the different polymers is rather complicated, because several factors are playing an important role on the final result: (i) the design of the electrochemical cell, (ii) the electrochemical method used for evaluation of the sensor, (iii) nature of the polymer, etc.
Only very few studies are published concerning the application of molecular imprinting technology for the analysis of SARS-CoV-2 proteins.There was a study describing the application of o-phenylenediamine o-phenylenediamine deposited on the macroporous gold screen-printed electrode with the receptor-binding domain of SARS-CoV-2-RBD for impedimetric measurements [59]. The described sensor was sensitive to the concentrations of SARS-CoV-2-RBD molecules in the range of pg/mL. In another study m-phenylenediamine (mPD) was imprinted with SARS-CoV-2 nucleoprotein (ncovNP). The sensitivity of the sensor according to the DPV signal was in the range of fM [58]. In order to demonstrate the selectivity of the sensor, BSA among others were used in the study. The Ppy was imprinted with gp51 and was applied in the design of electrochemical sensor [44]. The sensitivity of the sensor according to the results of simplified pulsed amperometric detection was in the range of μg/mL. The electrochemical sensors based on Ppy with imprints of prostate-specific antigen (PSA) was reported in 2020 [15]. The square wave voltammetry technique was used to determinate PSA concentration. The described sensor was sensitive to the concentrations of PSA molecules in the range of pg/mL. The electrochemical MIP sensor based on Ppy and aminophenylboronic acid (p-APBA) bilayer was imprinted with lysozyme [48]. The sensitivity of the sensor according to the CV signal was in the range of ppm. Hence, several factors govern the sensitivity of the MIP sensors. The electrochemical method of chronoamperometry (pulsed amperometric detection) by the sequence of potential pulses was only occasionally used in previous studies. In this study, we demonstrated that the obtained MIP-Ppy modified Pt electrodes can be applied for the determination of imprinted SARS-CoV-2-S glycoproteins.
Conclusions
Pt electrode was modified by two types of Ppy layers: (i) MIP-Ppy layer, which was modified by imprints of SARS-CoV-2-S glycoprotein and (ii) NIP-Ppy, which was formed without the imprint of any proteins. The comparison of the current changes on Pt electrode during the electrochemical deposition of MIP-Ppy and NIP-Ppy has demonstrated that the current for NIP-Ppy increased approximately by only 1.05 times more than that registered during the deposition of MIP-Ppy layer. This means that the SARS-CoV-2-S glycoprotein, which serves as the template molecule for MIP-Ppy layer, does not have a crucial effect on the thickness of the deposited polymer layer and the initial characteristics of the formed MIP-Ppy and NIP-Ppy layers are comparable. The comparison of calibration curves registered after the incubation of MIP-Ppy and NIP-Ppy modified Pt electrodes revealed that the interaction of SARS-CoV-2-S glycoprotein with MIP-Ppy generates 2.1 times higher change of current for MIP-Ppy modified electrode, in comparison with that registered for NIP-Ppy modified Pt electrode. The selectivity of SARS-CoV-2-S imprinted MIP-Ppy modified Pt electrode was tested in comparison to BSA solution. The obtained slope values during the evaluation of MIP-Ppy modified Pt electrode sensitivity towards BSA were significantly lower when compared with that towards SARS-CoV-2-S glycoprotein. The results of application of MIP-Ppy modified Pt electrodes demonstrated higher current changes and can be applied for selective determination of the imprinted SARS-CoV-2-S glycoprotein. Therefore, it can be concluded that the molecular imprinting of the conducting polymer might be applied for the development of the electrochemical sensor for the detection of SARS-CoV-2-S glycoprotein.
Supporting Material
Supporting material_AR.docx
CRediT authorship contribution statement
Vilma Ratautaite: Methodology, Investigation, Writing – original draft. Raimonda Boguzaite: Methodology, Investigation, Writing – original draft. Ernestas Brazys: Methodology, Investigation, Writing – original draft. Almira Ramanaviciene: . Evaldas Ciplys: . Mindaugas Juozapaitis: . Rimantas Slibinskas: . Mikhael Bechelany: . Arunas Ramanavicius: Supervision, Conceptualization, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This project has received funding from the Research Council of Lithuania (LMTLT), GILIBERT 2021 program agreement No S-LZ-21–4 and co-founded by Campus France grant No. 46593RA (PHC GILIBERT 2021).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.electacta.2021.139581.
Appendix. Supplementary materials
References
- 1.Shan B., Broza Y.Y., Li W., Wang Y., Wu S., Liu Z., Wang J., Gui S., Wang L., Zhang Z., Liu W., Zhou S., Jin W., Zhang Q., Hu D., Lin L., Zhang Q., Li W., Wang J., Liu H., Pan Y., Haick H. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano. 2020;14:12125–12132. doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- 2.Lamboll R.D., Jones C.D., Skeie R.B., Fiedler S., Samset B.H., Gillett N.P., Rogelj J., Forster P.M. Modifying emission scenario projections to account for the effects of COVID-19: protocol for COVID-MIP. Geoscientific model development discussions. 2020:1–20. (2020) [Google Scholar]
- 3.Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y.-.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D., Farrell M.J., Van Hooser S., Pandori M. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rockett R.J., Arnott A., Lam C., Sadsad R., Timms V., Gray K.-.A., Eden J.-.S., Chang S., Gall M., Draper J., Sim E.M., Bachmann N.L., Carter I., Basile K., Byun R., O'Sullivan M.V., Chen S.C.A., Maddocks S., Sorrell T.C., Dwyer D.E., Holmes E.C., Kok J., Prokopenko M., Sintchenko V. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat. Med. 2020;26:1398–1404. doi: 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- 8.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., Veinotte K., Egri S.B., Schaffner S.F., Lemieux J.E., Munro J.B., Rafique A., Barve A., Sabeti P.C., Kyratsous C.A., Dudkina N.V., Shen K., Luban J. Structural and functional analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e738. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Park Y.-.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calligari P., Bobone S., Ricci G., Bocedi A. Molecular investigation of SARS–CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y., Yang C., Xu X.-f., Xu W., Liu S.-w. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amanat F., Krammer F. SARS-CoV-2 Vaccines: status Report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu B., Guo H., Zhou P., Shi Z.-.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tretjakov A., Syritski V., Reut J., Boroznjak R., Öpik A. Molecularly imprinted polymer film interfaced with Surface Acoustic Wave technology as a sensing platform for label-free protein detection. Anal. Chim. Acta. 2016;902:182–188. doi: 10.1016/j.aca.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Mazouz Z., Mokni M., Fourati N., Zerrouki C., Barbault F., Seydou M., Kalfat R., Yaakoubi N., Omezzine A., Bouslema A., Othmane A. Computational approach and electrochemical measurements for protein detection with MIP-based sensor. Biosens. Bioelectron. 2020;151 doi: 10.1016/j.bios.2019.111978. [DOI] [PubMed] [Google Scholar]
- 16.Kidakova A., Boroznjak R., Reut J., Öpik A., Saarma M., Syritski V. Molecularly imprinted polymer-based SAW sensor for label-free detection of cerebral dopamine neurotrophic factor protein. Sens. Actuators B. 2020;308 [Google Scholar]
- 17.Thoelen R., Vansweevelt R., Duchateau J., Horemans F., D'Haen J., Lutsen L., Vanderzande D., Ameloot M., VandeVen M., Cleij T.J., Wagner P. A MIP-based impedimetric sensor for the detection of low-MW molecules. Biosens. Bioelectron. 2008;23:913–918. doi: 10.1016/j.bios.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Ramanavicius S., Ramanavicius A. Conducting Polymers in the Design of Biosensors and Biofuel Cells. Polymers (Basel) 2021;13:49. doi: 10.3390/polym13010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanavicius S., Jagminas A., Ramanavicius A. Advances in molecularly imprinted polymers based affinity sensors (Review) Polymers (Basel) 2021;13:974. doi: 10.3390/polym13060974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malitesta C., Mazzotta E., Picca R.A., Poma A., Chianella I., Piletsky S.A. MIP sensors - the electrochemical approach. Anal. Bioanal. Chem. 2012;402:1827–1846. doi: 10.1007/s00216-011-5405-5. [DOI] [PubMed] [Google Scholar]
- 21.Ayankojo A.G., Reut J., Ciocan V., Öpik A., Syritski V. Molecularly imprinted polymer-based sensor for electrochemical detection of erythromycin. Talanta. 2020;209 doi: 10.1016/j.talanta.2019.120502. [DOI] [PubMed] [Google Scholar]
- 22.Ansari S., Masoum S. Molecularly imprinted polymers for capturing and sensing proteins: current progress and future implications. TrAC Trends Anal. Chem. 2019;114:29–47. [Google Scholar]
- 23.El-Sharif H.F., Stevenson D., Reddy S.M. MIP-based protein profiling: a method for interspecies discrimination. Sens. Actuators B. 2017;241:33–39. [Google Scholar]
- 24.Ratautaite V., Janssens S.D., Haenen K., Nesládek M., Ramanaviciene A., Baleviciute I., Ramanavicius A. Molecularly imprinted polypyrrole based impedimentric sensor for theophylline determination. Electrochim. Acta. 2014;130:361–367. [Google Scholar]
- 25.Ndunda E.N. Molecularly imprinted polymers—A closer look at the control polymer used in determining the imprinting effect: a mini review. J. Mol. Recognit. 2020;33:e2855. doi: 10.1002/jmr.2855. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad O.S., Bedwell T.S., Esen C., Garcia-Cruz A., Piletsky S.A. Molecularly imprinted polymers in electrochemical and optical sensors. Trends Biotechnol. 2019;37:294–309. doi: 10.1016/j.tibtech.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Yang B., Fu C., Li J., Xu G. Frontiers in highly sensitive molecularly imprinted electrochemical sensors: challenges and strategies. TrAC Trends Anal. Chem. 2018;105:52–67. [Google Scholar]
- 28.Alizadeh T., Ganjali M.R., Zare M., Norouzi P. Development of a voltammetric sensor based on a molecularly imprinted polymer (MIP) for caffeine measurement. Electrochim. Acta. 2010;55:1568–1574. [Google Scholar]
- 29.Plausinaitis D., Sinkevicius L., Samukaite-Bubniene U., Ratautaite V., Ramanavicius A. Evaluation of electrochemical quartz crystal microbalance based sensor modified by uric acid-imprinted polypyrrole. Talanta. 2020;220 doi: 10.1016/j.talanta.2020.121414. [DOI] [PubMed] [Google Scholar]
- 30.Lowdon J.W., Diliën H., Singla P., Peeters M., Cleij T.J., van Grinsven B., Eersels K. MIPs for commercial application in low-cost sensors and assays – An overview of the current status quo. Sens. Actuators B. 2020;325 doi: 10.1016/j.snb.2020.128973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selvolini G., Marrazza G. MIP-based sensors: promising new tools for cancer biomarker determination. Sensors. 2017;17:718. doi: 10.3390/s17040718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratautaite V., Plausinaitis D., Baleviciute I., Mikoliunaite L., Ramanaviciene A., Ramanavicius A. Characterization of caffeine-imprinted polypyrrole by a quartz crystal microbalance and electrochemical impedance spectroscopy. Sens. Actuators B. 2015;212:63–71. [Google Scholar]
- 33.Ratautaite V., Nesladek M., Ramanaviciene A., Baleviciute I., Ramanavicius A. Evaluation of histamine imprinted polypyrrole deposited on boron doped nanocrystalline diamond. Electroanalysis. 2014;26:2458–2464. [Google Scholar]
- 34.Dabrowski M., Lach P., Cieplak M., Kutner W. Nanostructured molecularly imprinted polymers for protein chemosensing. Biosens. Bioelectron. 2018;102:17–26. doi: 10.1016/j.bios.2017.10.045. [DOI] [PubMed] [Google Scholar]
- 35.Tamboli V.K., Bhalla N., Jolly P., Bowen C.R., Taylor J.T., Bowen J.L., Allender C.J., Estrela P. Hybrid synthetic receptors on MOSFET devices for detection of prostate specific antigen in human plasma. Anal. Chem. 2016;88:11486–11490. doi: 10.1021/acs.analchem.6b02619. [DOI] [PubMed] [Google Scholar]
- 36.Zeng Q., Huang X., Ma M. A molecularly imprinted electrochemical sensor based on polypyrrole/carbon nanotubes composite for the detection of S-ovalbumin in egg white. Int. J. Electrochem. Sci. 2017;12:3965–3981. [Google Scholar]
- 37.Shumyantseva V.V., Bulko T.V., Sigolaeva L.V., Kuzikov A.V., Archakov A.I. Electrosynthesis and binding properties of molecularly imprinted poly-o-phenylenediamine for selective recognition and direct electrochemical detection of myoglobin. Biosens. Bioelectron. 2016;86:330–336. doi: 10.1016/j.bios.2016.05.101. [DOI] [PubMed] [Google Scholar]
- 38.Stojanovic Z., Erdőssy J., Keltai K., Scheller F.W., Gyurcsányi R.E. Electrosynthesized molecularly imprinted polyscopoletin nanofilms for human serum albumin detection. Anal. Chim. Acta. 2017;977:1–9. doi: 10.1016/j.aca.2017.04.043. [DOI] [PubMed] [Google Scholar]
- 39.Erdőssy J., Horváth V., Yarman A., Scheller F.W., Gyurcsányi R.E. Electrosynthesized molecularly imprinted polymers for protein recognition. TrAC Trends Anal. Chem. 2016;79:179–190. [Google Scholar]
- 40.Yarman A., Dechtrirat D., Bosserdt M., Jetzschmann K.J., Gajovic-Eichelmann N., Scheller F.W. Cytochrome c-derived hybrid systems based on moleculary imprinted polymers. Electroanalysis. 2015;27:573–586. [Google Scholar]
- 41.Qian L.-.W., Hu X.-.L., Guan P., Gao B., Wang D., Wang C.-.L., Li J., Du C.-.B., Song W.-.Q. Thermal preparation of lysozyme-imprinted microspheres by using ionic liquid as a stabilizer. Anal. Bioanal. Chem. 2014;406:7221–7231. doi: 10.1007/s00216-014-8133-9. [DOI] [PubMed] [Google Scholar]
- 42.Wu S., Tan W., Xu H. Protein molecularly imprinted polyacrylamide membrane: for hemoglobin sensing. Analyst. 2010;135:2523–2527. doi: 10.1039/c0an00191k. [DOI] [PubMed] [Google Scholar]
- 43.Tlili A., Attia G., Khaoulani S., Mazouz Z., Zerrouki C., Yaakoubi N., Othmane A., Fourati N. Contribution to the understanding of the interaction between a polydopamine molecular imprint and a protein model: ionic strength and ph effect investigation. Sensors. 2021;21:619. doi: 10.3390/s21020619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramanaviciene A., Ramanavicius A. Molecularly imprinted polypyrrole-based synthetic receptor for direct detection of bovine leukemia virus glycoproteins. Biosens. Bioelectron. 2004;20:1076–1082. doi: 10.1016/j.bios.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 45.Baleviciute I., Ratautaite V., Ramanaviciene A., Balevicius Z., Broeders J., Croux D., McDonald M., Vahidpour F., Thoelen R., Ceuninck W.D., Haenen K., Nesladek M., Reza A., Ramanavicius A. Evaluation of theophylline imprinted polypyrrole film. Synth. Met. 2015;209:206–211. [Google Scholar]
- 46.Ratautaite V., Topkaya S.N., Mikoliunaite L., Ozsoz M., Oztekin Y., Ramanaviciene A., Ramanavicius A. Molecularly Imprinted Polypyrrole for DNA Determination. Electroanalysis. 2013;25:1169–1177. [Google Scholar]
- 47.Viter R., Kunene K., Genys P., Jevdokimovs D., Erts D., Sutka A., Bisetty K., Viksna A., Ramanaviciene A., Ramanavicius A. Photoelectrochemical bisphenol S sensor based on ZnO-nanoroads modified by molecularly imprinted polypyrrole. Macromol. Chem. Phys., 2020;221 [Google Scholar]
- 48.Rick J., Chou T.-.C. Amperometric protein sensor – fabricated as a polypyrrole, poly-aminophenylboronic acid bilayer. Biosens. Bioelectron. 2006;22:329–335. doi: 10.1016/j.bios.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Kan X., Xing Z., Zhu A., Zhao Z., Xu G., Li C., Zhou H. Molecularly imprinted polymers based electrochemical sensor for bovine hemoglobin recognition. Sens. Actuators B. 2012;168:395–401. [Google Scholar]
- 50.Chen H.-.J., Zhang Z.-.H., Luo L.-.J., Yao S.-.Z. Surface-imprinted chitosan-coated magnetic nanoparticles modified multi-walled carbon nanotubes biosensor for detection of bovine serum albumin. Sens. Actuators B. 2012;163:76–83. [Google Scholar]
- 51.Yola M.L., Atar N. Development of cardiac troponin-I biosensor based on boron nitride quantum dots including molecularly imprinted polymer. Biosens. Bioelectron. 2019;126:418–424. doi: 10.1016/j.bios.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 52.Silva B.V.M., Rodríguez B.A.G., Sales G.F., Sotomayor M.D.P.T., Dutra R.F. An ultrasensitive human cardiac troponin T graphene screen-printed electrode based on electropolymerized-molecularly imprinted conducting polymer. Biosens. Bioelectron. 2016;77:978–985. doi: 10.1016/j.bios.2015.10.068. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z., Li F., Xia J., Xia L., Zhang F., Bi S., Shi G., Xia Y., Liu J., Li Y., Xia L. An ionic liquid-modified graphene based molecular imprinting electrochemical sensor for sensitive detection of bovine hemoglobin. Biosens. Bioelectron. 2014;61:391–396. doi: 10.1016/j.bios.2014.05.043. [DOI] [PubMed] [Google Scholar]
- 54.Li L., Yang L., Xing Z., Lu X., Kan X. Surface molecularly imprinted polymers-based electrochemical sensor for bovine hemoglobin recognition. Analyst. 2013;138:6962–6968. doi: 10.1039/c3an01435e. [DOI] [PubMed] [Google Scholar]
- 55.Ramanavicius S., Ramanavicius A. Charge transfer and biocompatibility aspects in conducting polymer-based enzymatic biosensors and biofuel cells. Nanomaterials. 2021;11:371. doi: 10.3390/nano11020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plausinaitis D., Ratautaite V., Mikoliunaite L., Sinkevicius L., Ramanaviciene A., Ramanavicius A. Quartz crystal microbalance-based evaluation of the electrochemical formation of an aggregated polypyrrole particle-based layer. Langmuir. 2015;31:3186–3193. doi: 10.1021/la504340u. [DOI] [PubMed] [Google Scholar]
- 57.Parisi O.I., Dattilo M., Patitucci F., Malivindi R., Pezzi V., Perrotta I., Ruffo M., Amone F., Puoci F. Monoclonal-type” plastic antibodies for SARS-CoV-2 based on molecularly imprinted polymers. bioRxiv. 2020 2020.2005.2028.120709. [Google Scholar]
- 58.Raziq A., Kidakova A., Boroznjak R., Reut J., Öpik A., Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tabrizi M.A., Fernández-Blázquez J.P., Medina D.M., Acedo P. An ultrasensitive molecularly imprinted polymer-based electrochemical sensor for the determination of SARS-CoV-2-RBD by using macroporous gold screen-printed electrode. Biosens. Bioelectron. 2021 doi: 10.1016/j.bios.2021.113729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramanavicius A., Oztekin Y., Ramanaviciene A. Electrochemical formation of polypyrrole-based layer for immunosensor design. Sens. Actuators B. 2014;197:237–243. [Google Scholar]
- 61.Svenson J., Andersson H.S., Piletsky S.A., Nicholls I.A. Spectroscopic studies of the molecular imprinting self-assembly process. J. Mol. Recognit. 1998;11:83–86. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<83::AID-JMR395>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 62.Ermis N., Tınkılıc N. Preparation of molecularly imprinted polypyrrole modified gold electrode for determination of tyrosine in biological samples. Int. J. Electrochem. Sci. 2018;13:2286–2298. [Google Scholar]
- 63.Heinze J., Frontana-Uribe B.A., Ludwigs S. Electrochemistry of conducting polymers—persistent models and new concepts. Chem. Rev. 2010;110:4724–4771. doi: 10.1021/cr900226k. [DOI] [PubMed] [Google Scholar]
- 64.Lewis T.W., Wallace G.G., Kim C.Y., Kim D.Y. Studies of the overoxidation of polypyrrole. Synth. Met. 1997;84:403–404. [Google Scholar]
- 65.de Wit J.N., Kessel T.v. Effects of ionic strength on the solubility of whey protein products. A colloid chemical approach. Food Hydrocoll. 1996;10:143–149. [Google Scholar]
- 66.Beauchamp D.L., Khajehpour M. Studying salt effects on protein stability using ribonuclease t1 as a model system. Biophys. Chem. 2012;161:29–38. doi: 10.1016/j.bpc.2011.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.