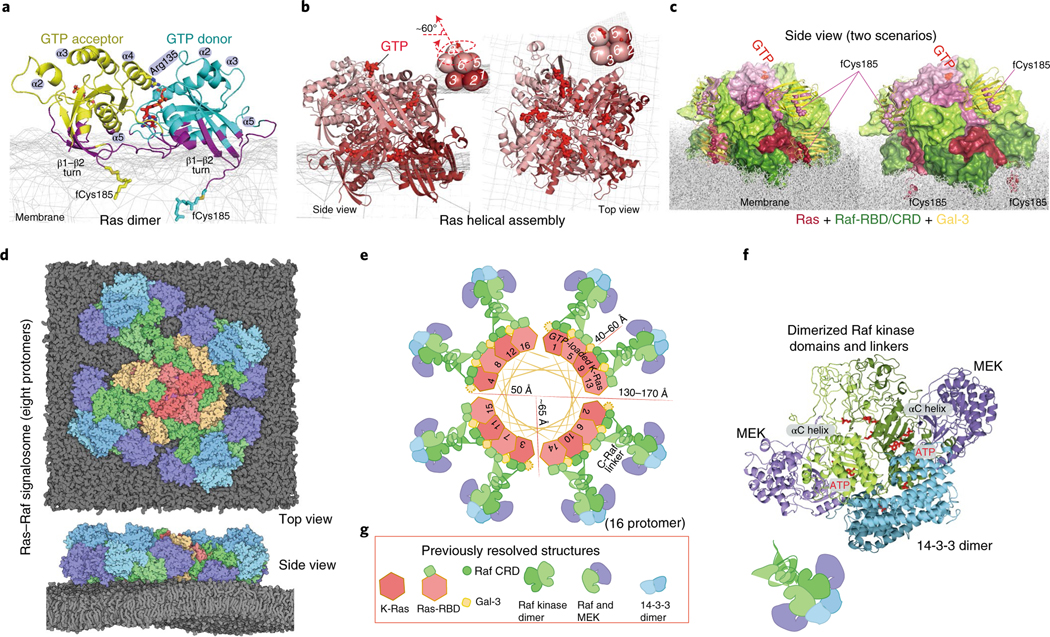
Fig. 1 |. Outline of the structural model of a Ras–Raf signalosome.
a, The GMA K-Ras dimer model. Residues contacting the membrane (mesh) are shown in purple; the GTP at the interface is in red. b, The (eight-protomer) K-Ras helical assembly with illustrative cartoons. c, The K-Ras helical assembly (pink), decorated with C-Raf RBD and CRD domains (green) and Gal-3 proteins (yellow). Two scenarios—one with Gal-3 at the base tier (left) and one without (right)—are shown, with the same orientation. d, The signalosome model on the membrane. The eight-protomer model contains four catalytic units at the periphery. e, Diagram of the signalosome model with a helical wheel representing the Ras assembly at the center. The Gal-3 molecules associated with the base-tier K-Ras (1–4) are outlined by dashed lines to indicate that Gal-3 is not structurally required at the base tier. To illustrate how the signalosome could grow beyond eight protomers, 16 protomers are shown. f, One catalytic unit centered on a Raf kinase dimer. ATP and phosphorylated Raf residues (pSer338, pTyr341 and pSer621) are in red. g, Previously resolved structures used as building blocks in the modeling: the K-Ras monomer85 (PDB 4DSN), H-Ras–Raf RBD complex47 (PDB 4G0N), C-Raf CRD86 (PDB 1FAR), Gal-3 (ref. 87) (PDB 3ZSM), C-Raf kinase domain dimer64 (PDB 3OMV), B-Raf–MEK1 complex65 (PDB 4MNE) and 14–3-3σ dimer bound to C-Raf phosphopeptide63 (PDB 4IEA).