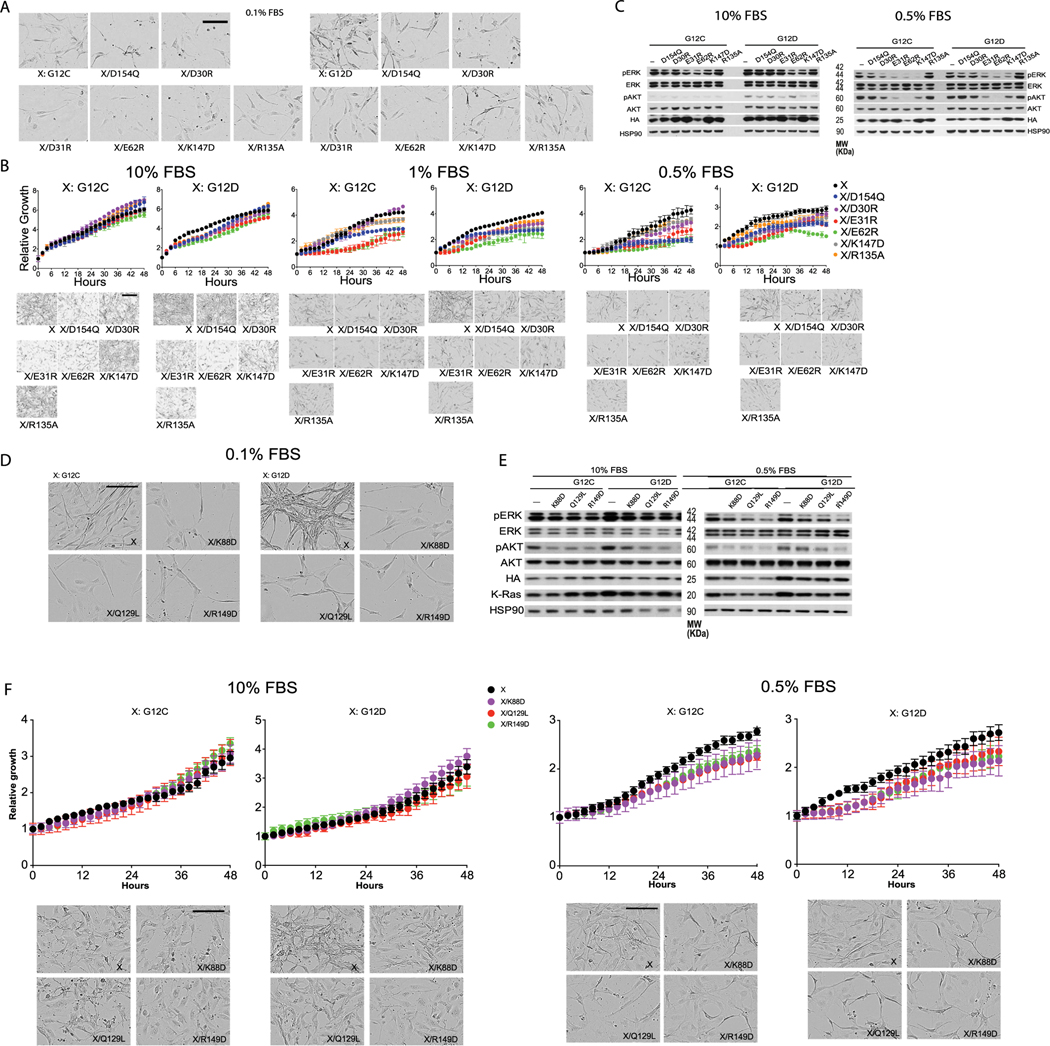
Extended Data Fig. 4 |. Cellular validation of the K-Ras signalosome interfaces.
Panels A–C here refer to the experiments testing mutations at the GMA dimer interface, and panels D–F refer to the experiments testing secondary/tertiary K-Ras interfaces. a. Representative cell images at the endpoint of the experiment testing mutations at the GMA dimer interface (Fig. 3f); scale bar: 50 μm. b. Growth rates of K-Raslox/K-RASMUT cells expressing the indicated mutations in cis with either G12C or G12D mutations in 10%, 1%, and 0.5% FBS medium, represented by the confluence value assessed by IncuCyte. Representative pictures at the endpoint are shown in the bottom panels (scale bar: 50 μm). Data are shown as mean +/− standard deviation (n = 3 biologically independent experiments). c. Phosphorylation of ERK and AKT in K-Raslox/K-RASMUT cells expressing the indicated mutations in cis with either G12C or G12D mutations. Cells were lysed after 48 hours incubation in 0.5% or 10% FBS, as indicated, and analyzed by Western blot. These results are representative of three independent experiments with similar results. d. Representative cell images at the endpoint of the experiment testing mutations at the secondary and tertiary K-Ras/K-Ras interfaces (Fig. 6b); scale bar: 50 μm. e. Phosphorylation of ERK and AKT in K-Raslox/K-RASMUT cells expressing the indicated mutations in cis with either G12C or G12D mutations. Cells were lysed after 48 hours incubation in 0.5% or 10% FBS as indicated and analyzed by Western blot. These results are representative of three independent experiments with similar results. f. Growth rates of K-Raslox/K-RASMUT cells expressing the indicated mutations in cis with either G12C or G12D mutations to test the secondary and tertiary K-Ras/K-Ras interfaces in the background of either G12C or G12D, shown as confluence values measured by IncuCyte. Representative images at the end point are also shown. Cells were kept in 0.5% or 10% FBS. Data are shown as mean +/− standard deviation (n = 3 biologically independent experiments).