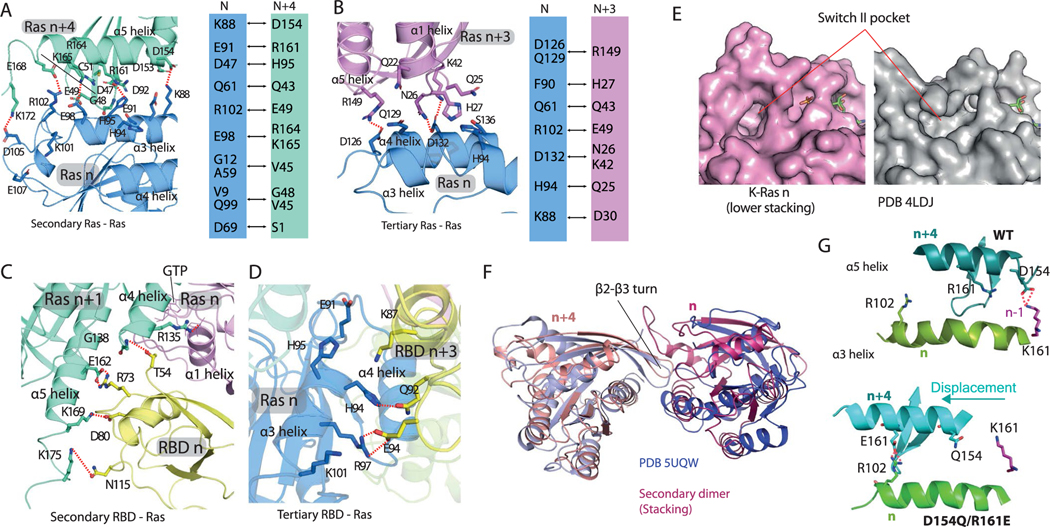
Extended Data Fig. 6 |. Structural details of secondary and tertiary Ras-Ras and Ras-RBD interactions.
a. Key contacts of the secondary (stacking) Ras-Ras interaction of K-Ras n (blue) and K-Ras n + 4 (green). A series of salt bridges form intermittently in MD simulations between the Lys88, Glu91, Glu98, Arg102, and Asp105 residues on the α3 helix of K-Ras n and the Asp154, Arg161, Lys165, Glu168, and Lys172 residues, respectively, on the α5 helix of K-Ras n + 4. b. Key contacts at the interface of K-Ras n + 3 (purple) and K-Ras n (blue). c. Key contacts of the secondary Ras-RBD interaction of C-Raf RBD n (yellow) with K-Ras n + 1 (green), which is the GTP acceptor of K-Ras n (purple), the primary binding partner of the RBD. The RBD also interacts with the HVR of K-Ras n + 1. Residues 1–53 of C-Raf are structurally unresolved but are likely to make additional interactions at this secondary Ras-RBD interface. d. Key contacts of the tertiary Ras-RBD interaction of Raf RBD n + 3 (yellow) and K-Ras n (blue). e. Unoccupied Switch-II pocket in a crystal structure of K-Ras (PDB 4LDJ) compared with the Switch-II pocket of K-Ras n in the stacking interaction. f. Superposition of stacking (n and n + 4) K-Ras proteins from the signalosome model with a crystal dimer of K-Ras (PDB 5UQW); in both cases the dimer interaction is mediated by a β2- β3 turn inserting into a Switch-II pocket. G. Comparison of D154Q/R161E with the WT in K-Ras oligomer simulations. As shown, the double mutation disrupts the D154-K147 salt bridge at the GMA dimer (n/n-1), and as compensation, allows an E161-R102 salt bridge in stacking (n/n + 4) by shifting the relative position of the α3 and α5 helices.