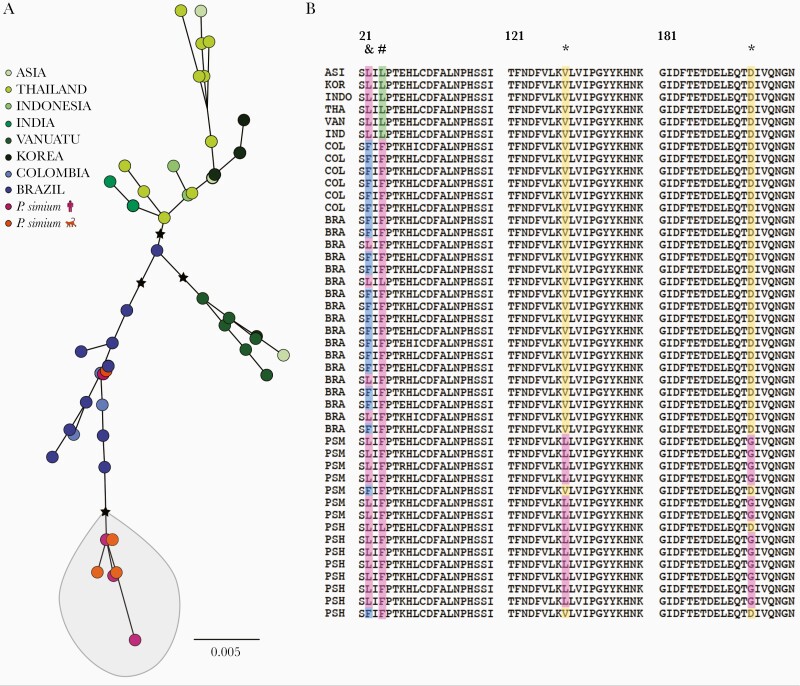
Figure 5.
Global diversity in the Plasmodium vivax gamete surface protein Pvs47, a key determinant of parasite-vector compatibility. (A) Maximum likelihood tree of Pvs47 protein sequences from P vivax and Plasmodium simium generated with the best-fit protein evolution model (FLU + I). Black stars indicate basal nodes with bootstrap support >85% (1000 pseudoreplicates). Note that P simium clusters with New World P vivax isolates. (B) Amino acid alignment of the variable domain of Pvs47 (residues 21–200); the full-length protein has 434 amino acids. * indicates amino acid changes private of P simium; # indicates an amino acid change shared between P simium and New World P vivax lineages; and & indicates an amino acid change private of New World P vivax lineages. A single Pvs47 sequence represents all Asian lineages of P vivax because they are >99% identical [34]. Sample origins are abbreviated as follows: ASI, Asia (country not specified); BRA, Amazonian Brazil; COL, Colombia; IND, India; INDO, Indonesia; KOR, South Korea; PSH, P simium of human origin; PSM, P simium of simian origin; THA, Thailand; and VAN, Vanuatu. Sample origins are given in Supplementary Table 3 and Pvs47 polymorphisms are listed in Supplementary Table 11.