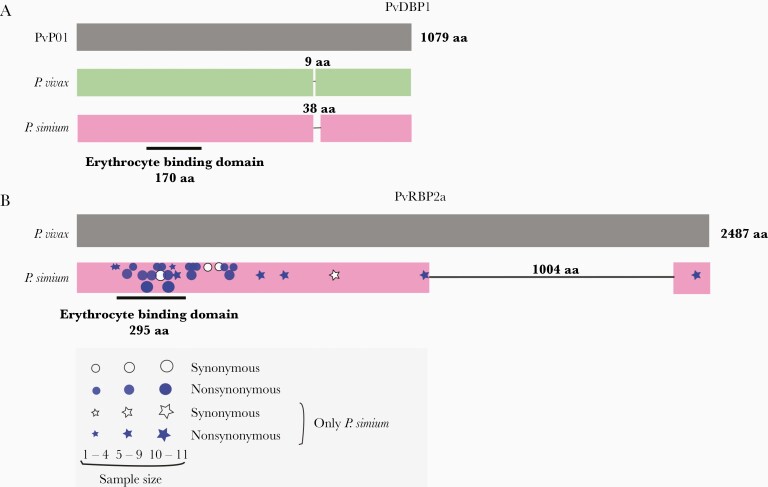
Figure 6.
Deletions in the Plasmodium simium PvDBP1 and PvRBP2a ligands of red blood cells. (A) Schematic representation of PvDBP1 showing the 9-amino acid (aa) deletion found in the vast majority of worldwide Plasmodium vivax isolates, relative to the PvP01 reference sequence from Papua New Guinea, and the 38-aa deletion that may occur in P simium isolates, but not in P vivax. The location of the 170-aa erythrocyte binding motif of PvDBP is indicated. (B) Schematic representation of PvRBP2a showing the 1004-aa deletion found in all P simium isolates but not in worldwide P vivax populations. The location of the 295-aa erythrocyte binding motif of PvRBP2a is indicated [43]. The approximate location of synonymous and nonsynonymous nucleotide replacements at the pvrbp2a locus observed in newly sequenced P simium genome sequences is indicated by white and blue symbols, respectively; symbol sizes are directly proportional to the number of sequences showing the mutation. Stars and circles indicate nucleotide replacements found only in P simium and those also found in P vivax populations, respectively. See Supplementary Figure 8 and Supplementary Table 11 for further details.