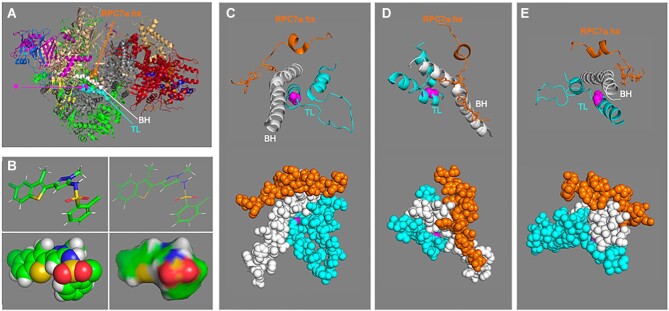
Figure 4.
Structure model of human apo-Pol III presumed amino acid target of inhibitor ML-60218 (ML6). Based on cryo-EM structure PDB 7D59 reported by Wang et al. (82), all viewed and produced with PyMOL Molecular Graphics system. (A) The features of interest are as follows: bridge helix (BH), and trigger loop (TL) regions of RPC1 are shown as spheres in white and cyan respectively while the RPC7α helix (hx) is in orange. This color coding is carried through in panels C-E. In this model the target glycine, G1045 of RPC1 is shown in magenta in sphere representation (indicated by arrow asterisk) and the rest of the 17-subunit model is in cartoon. RPC1 is colored grey; the color code follows that in figure 3B for the Pol III-specific subunits with the shared subunits in green. (B) Four representations of the inhibitor ML-60218 (C19H15Cl2N3O2S2, molecular mass: 452.4 Da (https://pubchem.ncbi.nlm.nih.gov/compound/RNA-Polymerase-III-Inhibitor). The top two are stick and line representations, and the bottom are sphere and surface, all in the same rotational view. (C−E) Zoomed in views of different rotations of the human Pol III structure in panel A after removal of other residues to better reveal features of the close-ups. The top parts show cartoon views as in A and the bottoms show the same view in sphere representation.