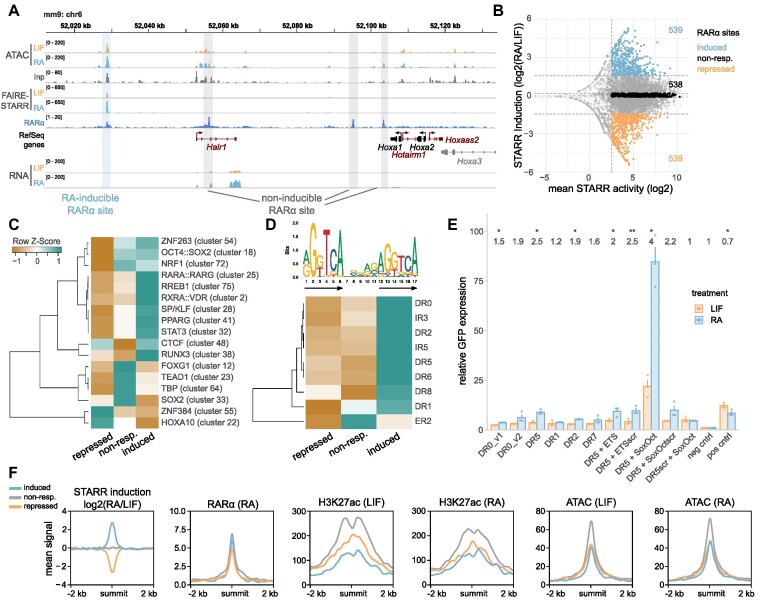
Figure 6.
RA-induced changes in enhancer activity at RARα-occupied sites correlate with specific sequence and chromatin features. (A) Genome browser view of an exemplary genomic region encompassing RA-inducible and non-inducible RARα binding sites. Normalized ATAC-, FAIRE-STARR and RNA-seq signals for LIF or RA treated cells and RefSeq genes for this region in either black (protein coding genes) or red (non-coding genes). (B) Distribution of changes in STARR activity (log2 fold change STARR score RA/LIF) and mean STARR activity (log score, for both treatments) of RARα-occupied regions that are covered in our FAIRE-STARR input library (as shown in Supplementary Figure S6A). Only regions with a minimum mean STARR activity ≥2.5 were included for further analysis. The 10% most induced, 10% most repressed and an equal number of regions that do not respond to RA treatment (non-resp.) were used for motif enrichment and TF binding analyses. (C) Enriched TF motif clusters (JASPAR 2018 clustered motif matrices) at induced, repressed, and non-responsive RARα-occupied sites. TF motif clusters with a maximum E-value of 1e−5 for at least one group and a log fold change ≥2 of induced or repressed over non-responsive regions are shown. Z-score normalization of E-values per row was performed. (D) Different spacings (0–8 nucleotides) and orientations (direct (DR), inverted (IR), and everted repeat (ER)) of the RARα::RXRα consensus motif (MA0159.1, upper panel, arrows highlight repeat orientation) were constructed in silico and used for motif enrichment analysis using AME. Only motifs which showed significant enrichment (E-value ≤ 1e−3) for at least one RARα binding site group are shown. Z-score normalization of E-values per row was performed. (E) Enhancer activity measured by STARR-RT-qPCR for selected spacing variants of the RARα::RXRα consensus motif (MA0159.1) and neighboring TF motifs as indicated (scr = scrambled motif) after 4 h of LIF or RA treatment. Bar plots depict the mean GFP expression + SE for three biological replicates. (F) Mean enrichment of RARα and H3K27ac as well as chromatin accessibility (ATAC) at induced, repressed, and non-responsive RARα-occupied sites.