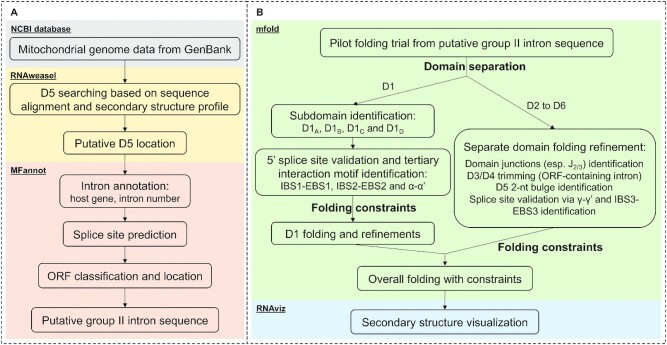
Figure 1.
Schematic of the overall workflow. (A) The group II intron discovery and annotation flow. Mitochondrial genome data from GenBank are subject to RNAweasel analysis to identify putative intron domain 5 location. The data are subsequently analyzed by MFannot to annotate the intron, predict intron splice sites and identify putative intronic opening reading frame (ORF). The outcome is the putative sequence for a given group II intron. (B) Secondary structure prediction workflow. The putative sequence for the group II intron is subject to folding prediction and refinement. In the pilot constraint-free folding trial, intron domains are identified and processed separately. Subsequent separate domain folding refinements yield folding constraints and validate the splice sites. Once folding constraints have been obtained from separate domain folding trials, the intron is folded in whole and the predicted secondary structure is subsequently visualized.