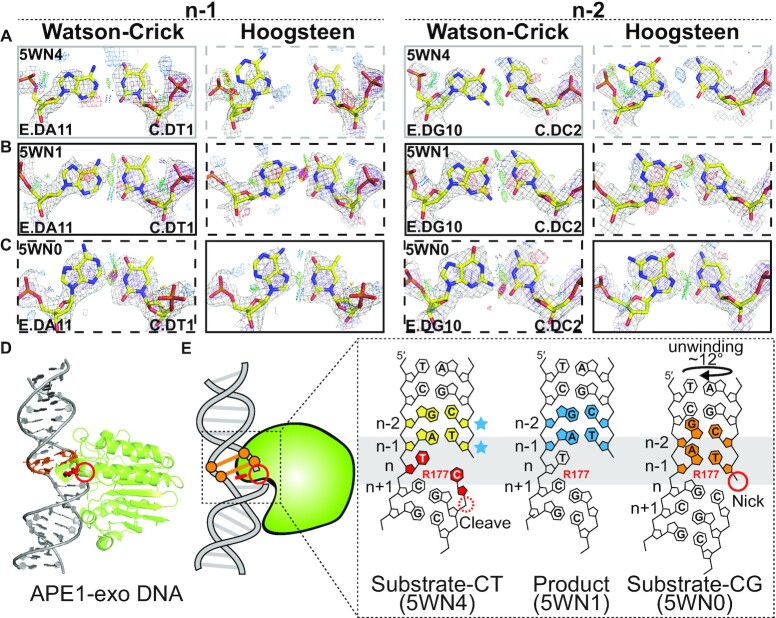
Figure 4.
Hoogsteen base pairs in APE1-exo. (A–C) Comparison of 2mFo-DFc and mFo-DFc electron density maps for the original Watson–Crick (left) and corresponding Hoogsteen models (right) for the A-T bp at position n-1 and G-C bp at position n-2 (A and B) with C-T mismatch at position n in (A) a substrate complex (PDB: 5WN4) and (B) a product complex (PDB: 5WN1), and (C) with C-G match at position n in a substrate complex (PDB: 5WN0). Electron density meshes and stereochemistry are as described in Figure 1C and the box scheme is as described in Figure 2A. (D) 3D structures of the protein–DNA complex showing the Hoogsteen bps. (E) Schematic showing the DNA containing Hoogsteen bps (in orange), Watson–Crick bps (in sky blue) and ambiguous Watson–Crick bps (in yellow next to sky blue stars), as well as the R177 and mismatches (in red). Also shown is the nick (red solid circle) and cleavage site (red dashed circle).