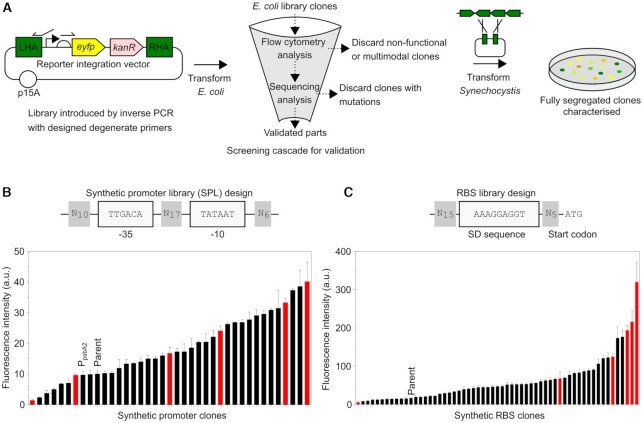
Figure 1.
Parts enabling rational and combinatorial pathway assembly in cyanobacteria. (A) Workflow of library design, generation and characterisation. Libraries were generated by inverse PCR and E. coli was transformed with each library. Clones were picked (70 randomly and 30 spanning a range of fluorescence levels by visual inspection) and a screening cascade of flow cytometry and sequencing was used for validation. Clones were discarded if their fluorescence was multimodal or weaker than pATM2 (−ve control) or if they contained unintended mutations (at non-degenerate positions). Synechocystis was transformed and integration and full segregation at the previously-used ndhB locus (ssl0410 CDS) neutral site was confirmed before characterisation. (B) Synthetic promoter library (SPL) design conserved −35 and −10 consensus sequences, and randomised the surrounding sequences. Synechocystis SPL clones were characterised by flow cytometry (detail in Supplementary Figure S8). A subset of six promoters was chosen (red bars). ‘Parent’ plasmid pATM2 was used as the template to generate the SPL. (C) RBS library design. The Shine-Dalgarno sequence was conserved and the surrounding sequences were randomised. Synechocystis RBS library clones were characterised by flow cytometry (detail in Supplementary Figure S9). A subset of six RBSs was chosen (red bars). ‘Parent’ plasmid pGT77 was used as the template to generate the RBS library. Error bars represent the standard deviation of three independent biological replicates.