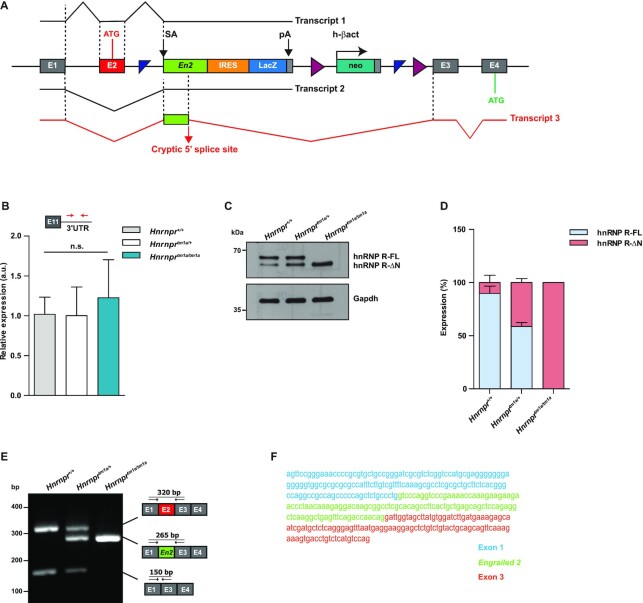
Figure 2.
A mouse model for knockout of the full-length isoform of Hnrnpr. (A) Schematic representation of the targeting strategy for disruption of the Hnrnpr locus. A gene-trap cassette containing a splice acceptor (SA) from the engrailed-2 (En2) gene, an internal ribosome entry site (IRES) directing the translation of a reporter (LacZ) and a SV40 polyadenylation sequence (pA) was inserted upstream of Hnrnpr exon 3. In Hnrnprtm1a/tm1a mice, splicing of exon 1 or exon 2 to En2 generates transcripts that terminate at the SV40 pA site (Transcripts 1 and 2). Utilization of a cryptic 5′ splice site in En2 results in a stable transcript (Transcript 3) with insertion of 115 nucleotides of En2 between exons 2 and 3 of Hnrnpr-ΔN mRNA. (B) qPCR analysis of total Hnrnpr (Hnrnpr-FL + Hnrnpr-ΔN) levels using primers specific to the 3′ UTR in brains of P5 Hnrnpr+/+,Hnrnprtm1a/+ and Hnrnprtm1a/tm1a mice. Data are mean ± SD (n = 4 animals). Statistical analysis was performed using one-way ANOVA followed by Tukey's multiple comparison test; n.s., not significant. (C) Western blot analysis of brain lysates obtained from P5 Hnrnpr+/+, Hnrnprtm1a/+ and Hnrnprtm1a/tm1a mice using an antibody against the C-terminal domain of hnRNP R. Gapdh was used as loading control. (D) Quantification of western blot data in (C). The expression of each isoform is presented as the percentage of total. Data are mean ± SD (n = 4 animals). (E) RT-PCR analysis of RNA from brains of P5 Hnrnpr+/+, Hnrnprtm1a/+ and Hnrnprtm1a/tm1a mice using primers within Hnrnpr exon 1 and 3. Schematic representations of observed amplicons are shown. (F) Sequence of the chimeric Hnrnpr-ΔN-En2 splice product detected in (E) containing exon 1, 115 nucleotides of En2 and Hnrnpr exon 3.