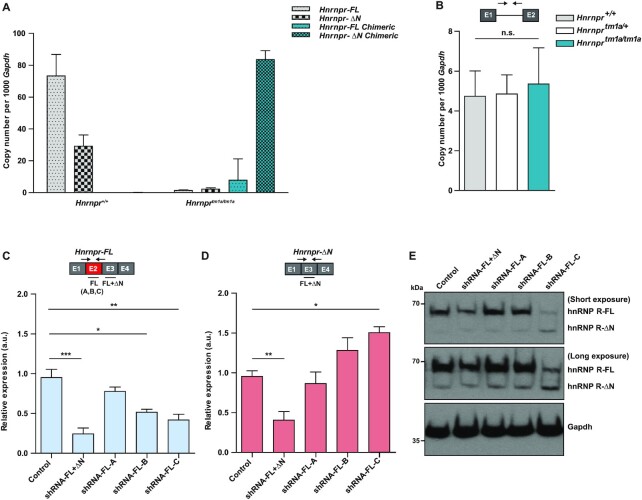
Figure 3.
Depletion of full-length hnRNP R results in an up-regulation of the hnRNP R-ΔN isoform. (A) Absolute mRNA copy numbers of wildtype and chimeric Hnrnpr isoforms normalized to copies of Gapdh in brains of Hnrnpr+/+,Hnrnprtm1a/+ and Hnrnprtm1a/tm1a mice. Data are mean ± SD (n = 4 animals). (B) qPCR analysis of Hnrnpr pre-mRNA levels using primers specific to intron 1 in brains of Hnrnpr+/+, Hnrnprtm1a/+ and Hnrnprtm1a/tm1a mice. Data are mean ± SD (n = 4 animals). Statistical analysis was performed using one-way ANOVA followed by Tukey's multiple comparison test; n.s., not significant. (C, D) qPCR analysis of Hnrnpr-FL (C) or Hnrnpr-ΔN (D) levels in primary motoneurons transduced with an shRNA against exon 3 for depletion of both hnRNP R isoforms (shRNA-FL+ΔN), or with different shRNAs against exon 2 for depletion of the full-length hnRNP R isoform (shRNA-FL-A, shRNA-FL-B and shRNA-FL-C). Absolute copy numbers of Hnrnpr-FL (C) and Hnrnpr-ΔN (D) mRNA isoforms were normalized to absolute copies of Gapdh, controls were set to 1. Data are mean ± SD (n = 4 independent experiments). Statistical analysis was performed using one-way ANOVA followed by Tukey's multiple comparison test; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (E) Western blot analysis of hnRNP R isoforms using a C-terminal-specific antibody performed on motoneurons transduced with control lentivirus or with lentiviruses for expression of the indicated shRNAs. Gapdh was used as loading control.