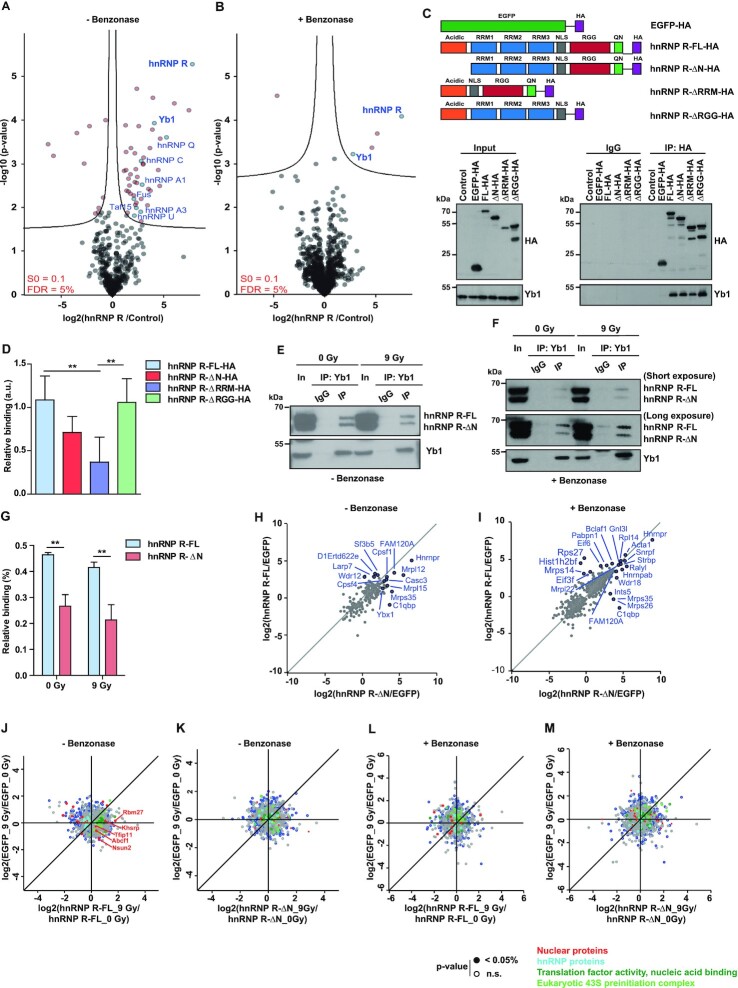
Figure 6.
Yb1 shows preferential interaction with the full-length isoform of hnRNP R. (A, B) Volcano plots showing proteins identified by mass spectrometry that were significantly enriched after immunoprecipitation with a C-terminal-specific hnRNP R antibody relative to IgG control immunoprecipitations. Experiments were carried out in the absence (A) or presence (B) of Benzonase. Log-transformed p-values (t test) associated with individual proteins were plotted against log-transformed fold changes in protein enrichment between hnRNP R and control immunoprecipitations. (C) Anti-HA immunoprecipitation from HEK293TN cells transfected with plasmids for expression of HA-tagged hnRNP R isoforms or deletion mutants, or HA-tagged EGFP, or from untransfected cells (control). Proteins were analyzed by western blot with antibodies against Yb1 and HA-tag. In, input; IP, immunoprecipitation. Input represents 5% of the lysate used for immunoprecipitation. (D) Quantification of Yb1 co-immunoprecipitation with hnRNP R constructs. Data are mean ± SD (n = 5 independent experiments). Statistical analysis was performed using two-way ANOVA followed by Bonferroni post-hoc test; **P ≤ 0.01. (E, F) Immunoprecipitation of Yb1 from lysates of non-irradiated (0 Gy) or irradiated (9 Gy) NSC-34 cells in the absence (E) or presence (F) of Benzonase treatment. Proteins were analyzed by Western blot with antibodies against Yb1 and hnRNP R. In, input; IP, immunoprecipitation. Input represents 5% of the lysate used for immunoprecipitation. (G) Quantification of hnRNP R isoforms co-immunoprecipitating with Yb1 in the presence of Benzonase (F) normalized to the input. Data are mean ± SD (n = 3 independent experiments). Statistical analysis was performed using two-way ANOVA followed by Bonferroni post-hoc test; **P ≤ 0.01. (H, I) Scatter plots showing proteins identified by mass spectrometry after immunoprecipitation with anti-HA antibody from HEK293TN transfected with plasmids for expression of hnRNP R-FL-HA, hnRNP R-ΔN-HA or EGFP-HA as control. Data are shown as log-transformed fold changes in protein enrichment between hnRNP R-FL-HA or hnRNP R-ΔN-HA and EGFP-HA immunoprecipitations, carried out in the absence (H) or presence (I) of Benzonase. (J, K) Scatter plots showing proteins identified by mass spectrometry after immunoprecipitation with anti-HA antibody from HEK293TN expressing hnRNP R-FL-HA (J), hnRNP R-ΔN-HA (K) or EGFP-HA under non-irradiated (0 Gy) or after exposure to irradiation (9 Gy). Experiments were carried out in the absence of Benzonase. Log-transformed fold changes in protein enrichment in EGFP-HA immunoprecipitations under non-irradiated (0 Gy) and irradiated (9 Gy) conditions were plotted against log-transformed fold changes in protein enrichment in hnRNP R-FL-HA (J) or hnRNP R-ΔN-HA (K) immunoprecipitations under non-irradiated (0 Gy) and irradiated (9 Gy) conditions. Functional annotations of proteins are colour-coded. (L, M) Same as (J, K) but in the presence of Benzonase.