Fig. 6.
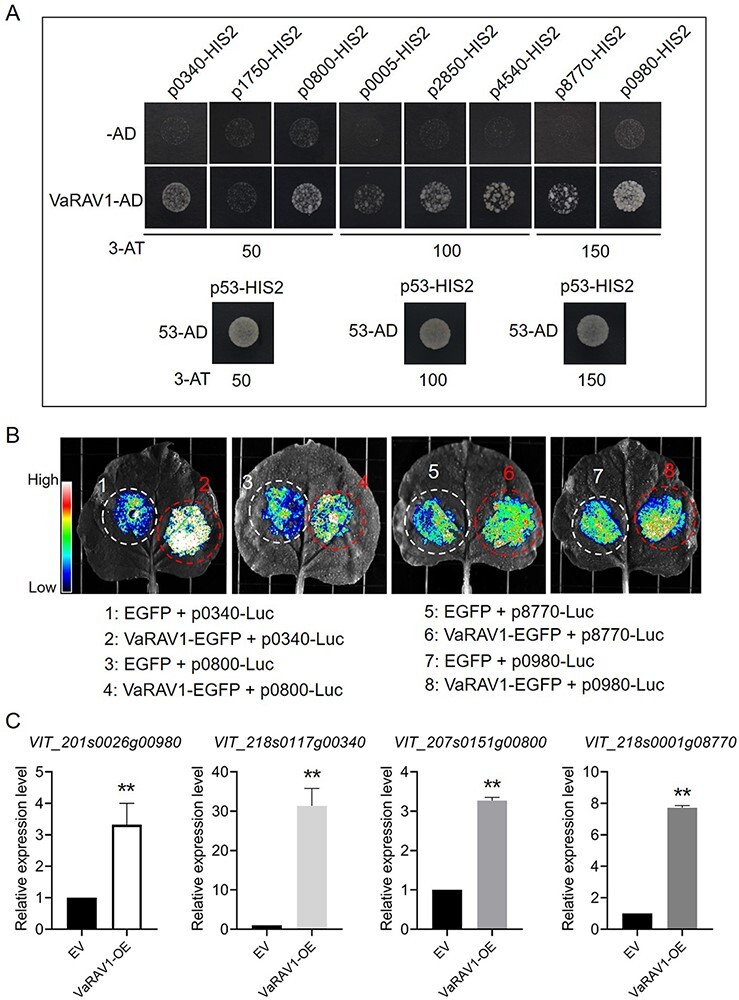
Detection of interactions between VaRAV1 and its target genes. (A) Yeast one-hybrid (Y1H) assay. The DNA sequences containing the predicted RAV1 motifs were synthesized and ligated into the bait vector pHIS2. The bait vectors and the prey vector harboring VaRAV1 fused to the GAL4 activation domain were introduced into the yeast strain Y187. Growth of transformed yeast cells was analyzed on SD/-Trp/-Leu/-His medium supplemented with 3-amino-1,2,4-triazole (3-AT). The p53HIS2 and pGADT-Rec2-53 were co-transformed into Y187 as positive control. For convenience, the last four numbers of gene IDs (Supplementary Table S3) were adopted for short. (B) Transient luciferase (Luc) activity assays. The promoters of the tested genes (VIT_218s0117g00340, VIT_207s0151g00800, VIT_218s0001g08770 and VIT_201s0026g00980) were ligated to the pCAMBIA1302-Luc reporter vector. Agrobacterium cells containing the Luc reporter vectors and p35S::VaRAV1-EGFP construct were co-infiltrated into N. benthamiana leaves. The possible interactions between VaRAV1 and tested genes were evaluated by Luc fluorescence intensities. (C) The expression profiles of VIT_218s0117g00340, VIT_207s0151g00800, VIT_218s0001g08770 and VIT_201s0026g00980 in the VaRAV1-OE cells. The expression of the four genes in the OE cells relative to the EV was measured by qPCR. Significant difference was determined by Student’s t-test. **P < 0.01.
