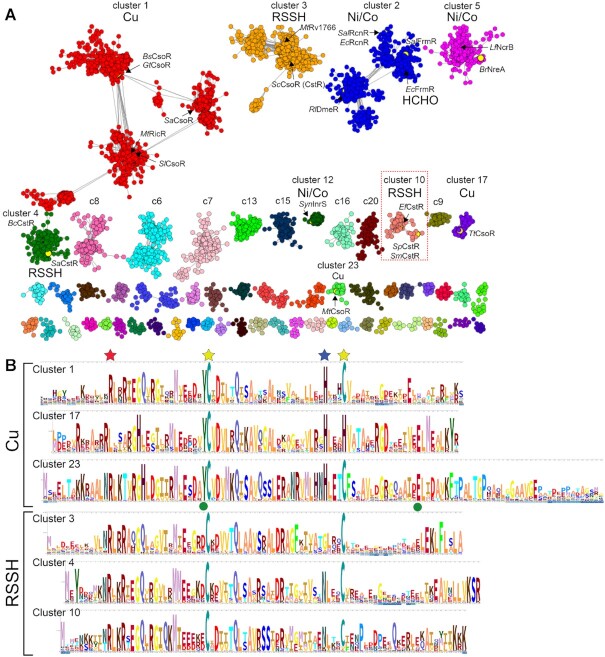
Figure 2.
Sequence similarity network (SSN) analysis of the CsoR superfamily of bacterial repressors. (A) Results of an SSN clustering analysis of 32,476 unique sequences belonging to the Pfam PF02583 using genomic enzymology tools and visualized using Cytoscape. Clusters are designated by a number and ranked according to the number of unique sequences (Supplementary Tables S1A, S2), color-coded, and functionally annotated as copper (Cu), nickel/cobalt (Ni/Co), persulfide (RSSH) or formaldehyde (HCHO) sensors, if known. Each node corresponds to sequences that are 80% identical over 80% of the sequence, using an alignment score of 28 (see Materials and Methods). Functionally characterized members in each of nine clusters (1–5, 10, 12, 17, 23) are indicated with species name and trivial name (25). Cluster 10 proteins are the subject of the work presented here. (B) Sequence logo representations of sequence conservation of the Cu(I) and RSSH sensors defined by the indicated cluster of sequences derived from panel A. The residues that coordinate the Cu(I) in CsoRs are marked by the yellow and blue stars, the residue equivalent to C9 in SpCstR (cluster 10) marked by a red star, and the two allosterically important second Cu(I) coordination shell residues in MtCsoR (cluster 23) (86) marked with a green circle. WebLogos for the Ni/Co and other uncharacterized PF02583 sensor clusters are also provided (Supplementary Figure S2A).