Figure 4.
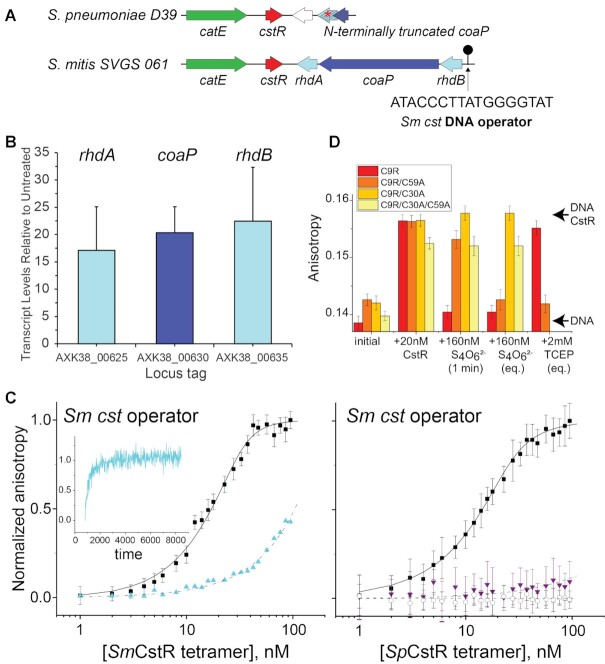
S. pneumoniae (Sp) D39 SPD_0073 encodes a bona fide persulfide-sensing CstR. (A) Genomic neighborhood and location of a candidate CstR-regulated operator-promoter site around selected streptococcal cstR genes (cluster 10; Figure 1A) investigated here. The DNA sequence of the core Sm cst operator is shown (30). (B) Induction of the expression of genes in S. mitis SVGS_061 by exogenous sulfide, as measured by qRT-PCR. AXK38_00625 (rhdA; see panel A); AXK38_00630 (coaP); AXK38_00635 (rhdB). (C) Left, SmCstR (locus tag AXK38_00620) binds to the S. mitis SVGS061 cst operator-containing DNA (black circles) and is inhibited by pre-treatment with CysSSH (cyan triangles, >90% persulfidated species according to the data in Figure 8) which is reversible upon addition of TCEP (cyan time course, inset). Right, representative DNA binding curves obtained for reduced (black circles) or tetrathionate-treated (purple triangles, >90% symmetric disulfide species according to the data in Supplementary Figure S9), C9A SpCstR on the S. mitis SVGS061 cst operator-containing DNA. Black open circles, C9A SpCstR binding to an AdcR operator (87), representative of non-specific binding. The continuous lines drawn through each data set represent the results of a fit to a two-tetramer binding model (Supplementary Figure S4B) (30), with parameters complied in Table 1. (D) Bar chart representation of the kinetics of Sm cst operator binding, dissociation by tetrathionate and subsequent reduction by TCEP by various SpCstR dithiol site mutants in a C9R SpCstR parent background, from left to right. The arrows labeled DNA CstR and DNA are the anisotropies associated with the CstR-bound and free DNAs. 1 min, anisotropy 1 min after addition of tetrathionate; eq, equilibrium anisotropy value.