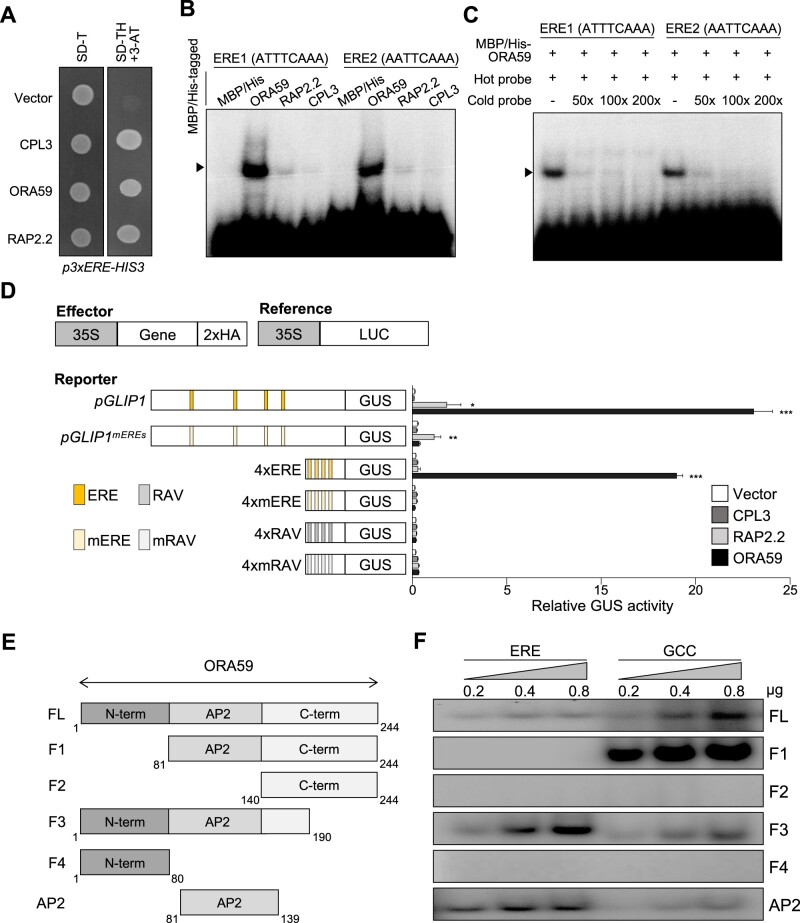
Figure 2.
ORA59 is the specific ERE-binding transcription factor. A, Isolation of ERE-binding transcription factors by Y1H screening. Yeast cells harboring the 3xERE-HIS3 reporter gene were transformed with effector constructs CPL3, ORA59, and RAP2.2. Transcription factor binding to ERE was tested on a selective medium lacking tryptophan and histidine, and supplemented with 0.2 mM 3-amino-1,2,4-triazole (SD-TH + 3-AT). B, DNA binding of ORA59, RAP2.2, and CPL3 to the two ERE sequences ATTTCAAA (ERE1) and AATTCAAA (ERE2). Recombinant proteins were incubated with biotin-labeled ERE oligonucleotide probes in EMSA. C, Competition assays. ORA59 binding to ERE1/2 was competed with increasing amounts (50×, 100×, 200×) of unlabeled oligonucleotide competitors. D, Transactivation analysis showing the ORA59-mediated GUS reporter gene induction driven by the GLIP1 and 4xERE synthetic promoters. The left part illustrates reference, effector, and reporter constructs. Reporter DNAs, either alone or together with effector DNAs, were transfected into protoplasts, and GUS activity was measured. Luciferase (LUC) expressed under control of the CaMV 35S promoter was used as an internal control (reference). Values represent means ± sd (n = 3 biological replicates). Asterisks indicate significant differences from vector control as determined by one-way ANOVA with Tukey test (*P < 0.05; **P < 0.01; ***P < 0.001). 35S, CaMV 35S; 2xHA, two copies of the HA tag sequence. E, Schematic diagram of full-length and truncated ORA59 proteins prepared for EMSA. F, DNA binding of different forms of ORA59 to the ERE and GCC box elements. Increasing amounts (0.2, 0.4, and 0.8 μg) of recombinant proteins were incubated with biotin-labeled ERE and GCC box oligonucleotide probes in EMSA. FL, full-length; GCC, GCC box.