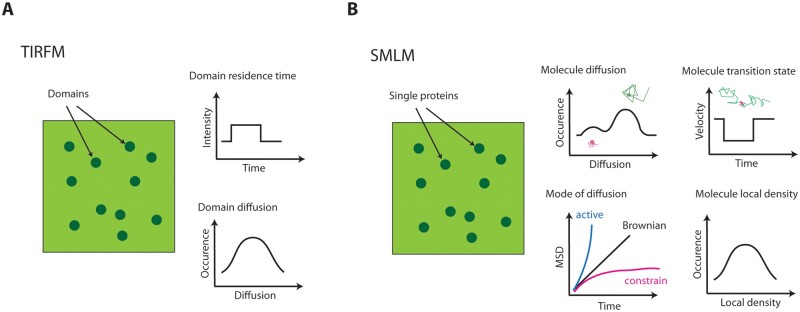
Figure 1.
Comparison of the type of data obtained with total internal reflexion fluorescence microscopy (TIRFM) and SMLM techniques. A, Under TIRFM, the number of fluorescent proteins in a given fluorescence-emitting domain cannot be determine experimentally. The duration of the signal can be related to the domain on–off at the PM (graph titled domain residence time). Domain displacement can also be documented (graph titled domain diffusion) and informs about the domain spatial dynamics. B, With SMLM, each single fluorescent dot corresponds to an individual emitter. Its individual diffusion can be estimated and may reflect some heterogeneity since molecules with high diffusion can co-exist with molecules of lower diffusion (graph titled molecule diffusion). MSD plot informs about the diffusion mode that could be either normal (Brownian) or abnormal (constrained or active; graph titled mode of diffusion). SMLM can also be used to determine molecule transition state. Typically, plot of velocity would inform about the molecule displacement for each time point of the track and reveals change of diffusion behavior along time (graph titled molecule transition state). This is particularly valuable to study protein recruitment in membrane nanodomains. To some extent, SMLM can also be used to study protein spatial organization. Localization of molecules along time can serve to calculate their local density (graph titled molecule local density), which can reveal where proteins form clusters and can be compared to TIRF observations