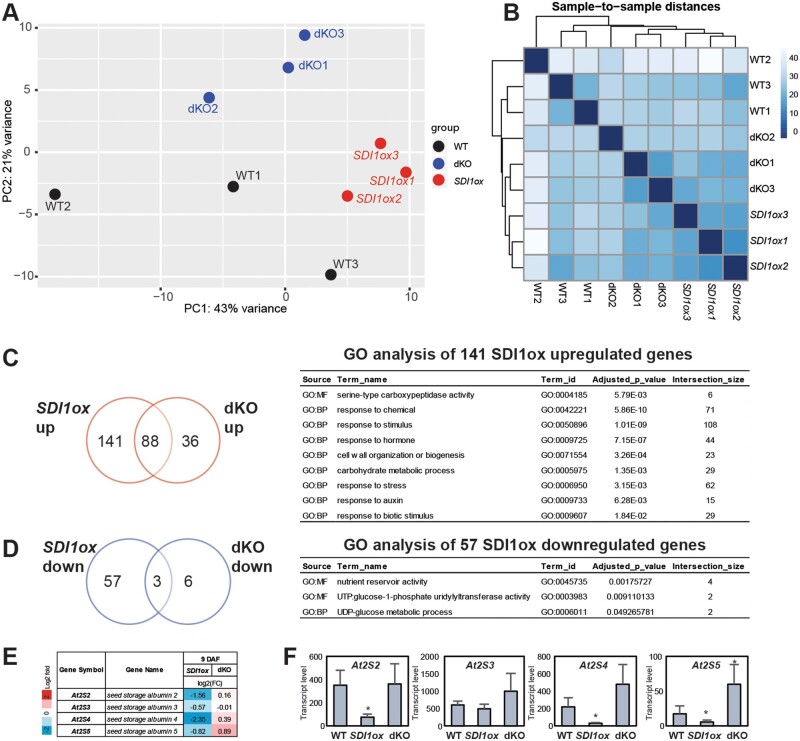
Figure 2.
Differential effects of SDI1 overexpression and sdi1sdi2 knockout on the seed transcriptome at 9 DAF. A, PCA plot explains the variance in normalized read counts from three biological sample libraries of, SDI1ox, dKO, and WT at 9 DAF along PC1 or X-axis and PC2 or Y-axis. B, Hierarchical clustering of the heatmap of the sample-to-sample distances demonstrates similarities and dissimilarities between samples. Venn diagrams of the significantly up- and downregulated genes in seeds of ox and dKO lines versus WTs depicted in red (C) and blue (D), respectively (selection of 1.5-fold threshold, FDR < 0.05) and the GO terms enriched for the SDI1ox-specific up/downregulated genes are depicted. Transcript profile of the genes encoding 2S-SSPs, is depicted in (E) and (F) quantified by RNA-seq and qRT-PCR, respectively. Data in F are mean ± sd of six replicates (three biological, two technical). Asterisks demonstrate significant changes, t test, P < 0.05. Error bars indicate standard deviation.