Figure 3.
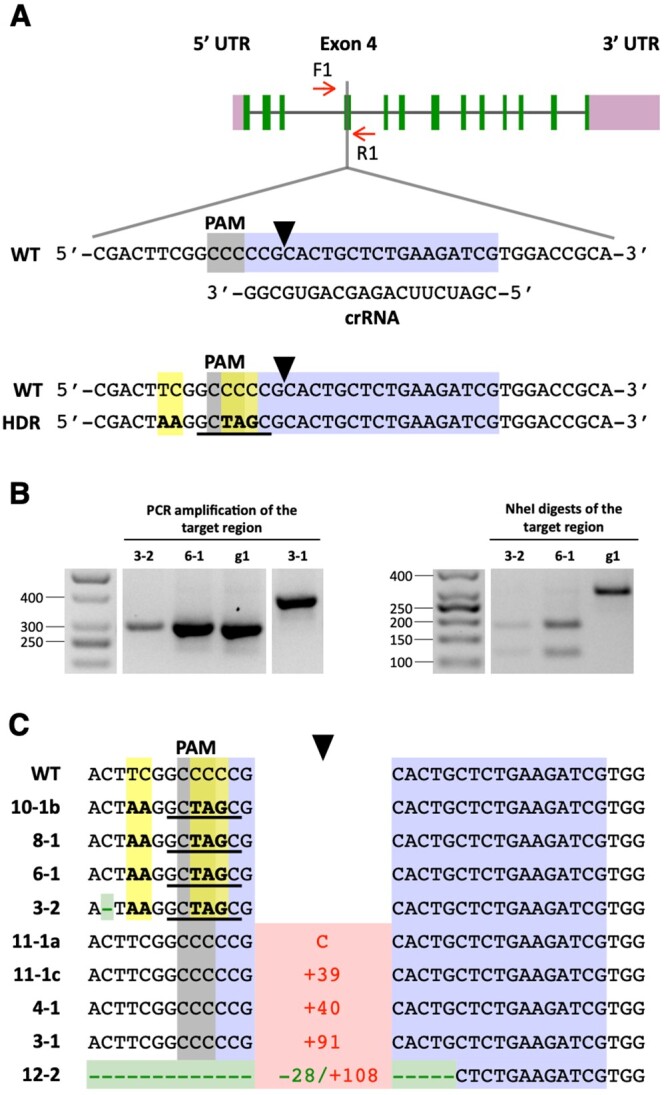
Co-editing of the FTSY gene in colonies selected for precise PPX1 editing. A, Schematic of the FTSY target region in exon 4. Color schemes and symbols are as described under Figure 1A. HDR of the DSB, using as template the transfected FTSY ssODN, is expected to introduce five base pair changes (highlighted in yellow) into the genome (bottom sequence). These sequence changes insert two stop codons (i.e. TAA and TAG) in the coding frame, destroy the PAM site, and create a new NheI restriction enzyme site (underlined in black). B, The FTSY target region of selected oxyfluorfen-resistant, pale green colonies was amplified by PCR (with primers F1 and R1) and the PCR products digested with NheI. The panels show representative reverse images of agarose resolved PCR products stained with ethidium bromide. The sizes of molecular weight markers are indicated in base pairs. g1, WT strain. C, DNA sequences of oxyfluorfen-resistant, pale green colonies, indicating alterations at the FTSY target site relative to the WT. Insertions, indicating type (C, cytosine) or number of base pairs, are depicted in red. Deletions, indicating type or number of base pairs, are depicted in green. Base substitutions are highlighted in yellow. Complete sequences for colonies exhibiting FTSY indels are shown in Supplemental Figure S5.
