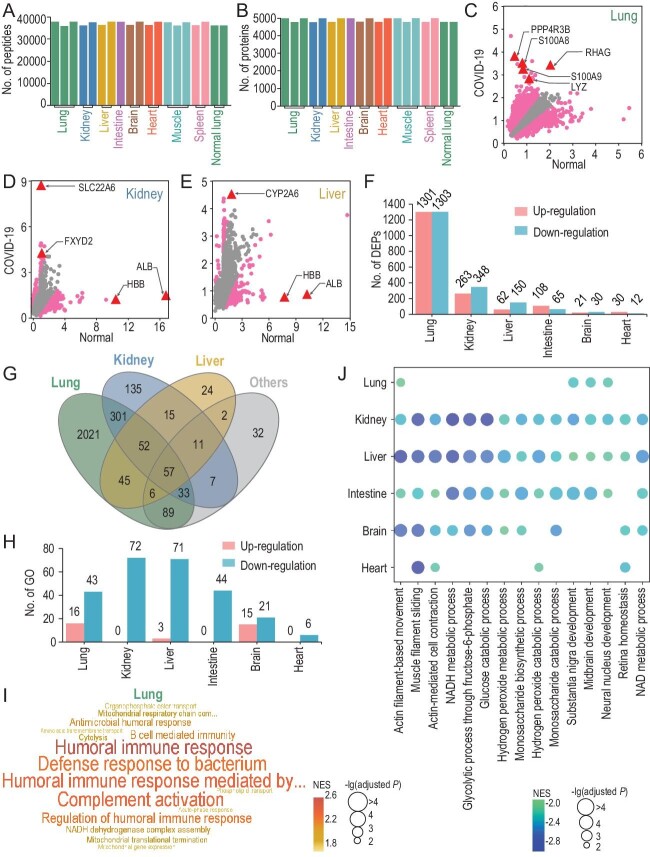
Figure 1.
The multi-omic study of COVID-19 post-mortem tissue samples. (A and B) The distribution of numbers of quantified (A) peptides and (B) proteins in 17 post-mortem and 2 normal lung tissues. (C–E) MAP-based identification of potential DEPs in the (C) lung, (D) kidney and (E) liver of post-mortem tissues. Arrows indicate the DEPs in each tissue are also shown as TSPs. (F) The distribution of numbers of up- and down-regulated DEPs in the six types of post-mortem tissues. (G) The overlap of DEPs in different post-mortem tissues. (H) The distribution of numbers of significantly up- or down-regulated Gene Ontology (GO) biological processes detected by GSEA (adjusted P-value < 0.01) in the six types of post-mortem tissues. (I) Visualization of up-regulated processes in post-mortem lungs, using a word cloud illustrator WocEA. (J) Down-regulated processes in ≥4 post-mortem tissue types.